| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,009,570 – 1,009,680 |
| Length | 110 |
| Max. P | 0.879842 |

| Location | 1,009,570 – 1,009,680 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 88.74 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -21.17 |
| Energy contribution | -21.18 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1009570 110 + 22224390 UUCUCCGAUUCUCCUUUUCUCUCCCACCCACACGUCUAACAUG-UAGAUAAAUCUGCUCGCGUAUUUCUCGCAGCGAUUUGCCAUAGCUCAGGCAGAGAGAUAGAUGGAGA .........(((((...((((((..........(((((.....-)))))(((((.(((.(((.......)))))))))))(((........))).)))))).....))))) ( -30.90) >DroSec_CAF1 95037 103 + 1 -----CGAUUCUCCUUUU--CUCCCGCCCACCCAUCUAACAUG-UAGAUAAAUCUGCUCGCGUAUUUUUCGCAGCGAUUUGCCAUAGCUCAUGCAGAGAGAUAGAUGGAGA -----....(((((.(((--(((..((......(((((.....-)))))(((((.(((.(((.......)))))))))))))....((....)).)))))).....))))) ( -25.80) >DroSim_CAF1 72230 108 + 1 UUCUCCGAUUCUCCUUUU--CUCCCGCCCACCCAUCUAACAUG-UAGAUAAAUCUGCUCGCGUAUUUUUCGCAGCGAUUUGCCAUAGCUCAUGCAGAGAGAUAGAUGGAGA .((((((..(((...(((--(((..((......(((((.....-)))))(((((.(((.(((.......)))))))))))))....((....)).)))))).))))))))) ( -27.80) >DroEre_CAF1 106331 104 + 1 UUCUCCAUUUCUCCGUUU--CUCC----CACCCC-UGGACAUGUUAGAUAAAUCUGCUCGCGUAUUUCUCGCAGCGAUUUGCCAUAGCUCAUGCAGAGAGAUAGAUGGAGA .(((((((((....((((--((((----((....-)))....((((..((((((.(((.(((.......))))))))))))...)))).......)))))))))))))))) ( -31.50) >consensus UUCUCCGAUUCUCCUUUU__CUCCCGCCCACCCAUCUAACAUG_UAGAUAAAUCUGCUCGCGUAUUUCUCGCAGCGAUUUGCCAUAGCUCAUGCAGAGAGAUAGAUGGAGA .........(((((......(((.......................(.((((((.(((.(((.......)))))))))))).)...((....))...)))......))))) (-21.17 = -21.18 + 0.00)

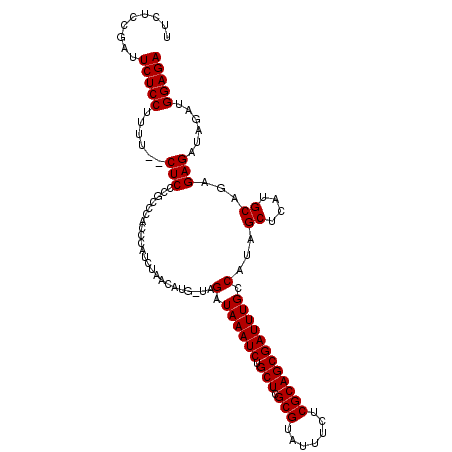
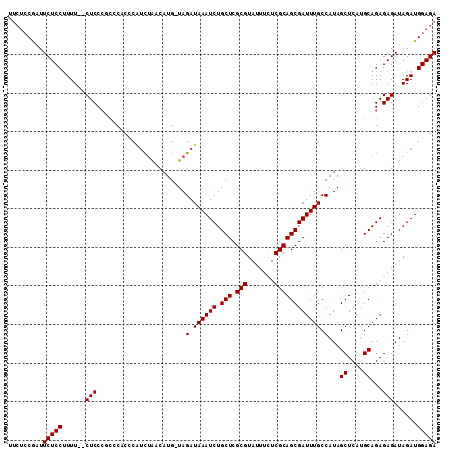
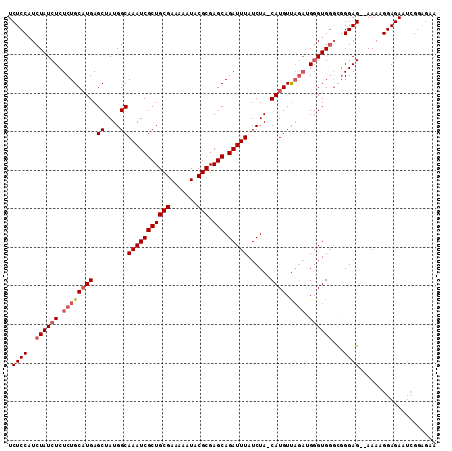
| Location | 1,009,570 – 1,009,680 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 88.74 |
| Mean single sequence MFE | -35.28 |
| Consensus MFE | -25.01 |
| Energy contribution | -26.32 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1009570 110 - 22224390 UCUCCAUCUAUCUCUCUGCCUGAGCUAUGGCAAAUCGCUGCGAGAAAUACGCGAGCAGAUUUAUCUA-CAUGUUAGACGUGUGGGUGGGAGAGAAAAGGAGAAUCGGAGAA (((((.(((.((((((((((........)))((((((((((((....).))).))).)))))(((((-((((.....))))))))).))))))).....)))...))))). ( -41.10) >DroSec_CAF1 95037 103 - 1 UCUCCAUCUAUCUCUCUGCAUGAGCUAUGGCAAAUCGCUGCGAAAAAUACGCGAGCAGAUUUAUCUA-CAUGUUAGAUGGGUGGGCGGGAG--AAAAGGAGAAUCG----- (((((.(((((((.((((((((.((....))(((((((((((.......))).))).))))).....-)))).)))).)))))))..))))--)............----- ( -33.20) >DroSim_CAF1 72230 108 - 1 UCUCCAUCUAUCUCUCUGCAUGAGCUAUGGCAAAUCGCUGCGAAAAAUACGCGAGCAGAUUUAUCUA-CAUGUUAGAUGGGUGGGCGGGAG--AAAAGGAGAAUCGGAGAA (((((.(((((((.((((((((.((....))(((((((((((.......))).))).))))).....-)))).)))).)))))))..))))--)................. ( -33.20) >DroEre_CAF1 106331 104 - 1 UCUCCAUCUAUCUCUCUGCAUGAGCUAUGGCAAAUCGCUGCGAGAAAUACGCGAGCAGAUUUAUCUAACAUGUCCA-GGGGUG----GGAG--AAACGGAGAAAUGGAGAA (((((((((((((((..(((((.((....))((((((((((((....).))).))).)))))......)))))..)-))))))----)...--..........))))))). ( -33.61) >consensus UCUCCAUCUAUCUCUCUGCAUGAGCUAUGGCAAAUCGCUGCGAAAAAUACGCGAGCAGAUUUAUCUA_CAUGUUAGAUGGGUGGGCGGGAG__AAAAGGAGAAUCGGAGAA .((((..((((((.((((((((.((....))(((((((((((.......))).))).)))))......)))).)))).))))))...)))).................... (-25.01 = -26.32 + 1.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:38 2006