
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,466,717 – 8,466,934 |
| Length | 217 |
| Max. P | 0.704721 |

| Location | 8,466,717 – 8,466,820 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.16 |
| Mean single sequence MFE | -29.05 |
| Consensus MFE | -21.20 |
| Energy contribution | -21.61 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8466717 103 + 22224390 ---------U-CUUCAGCUGAAAAUGUCGUUGGCU------GCCAGUUAAAGUUGGUGACGAGGCGUGAG-AGGUCUUUGUCACUACUGUUUUGCUUUGUGUUUCGGCUUUUGGCAUUAG ---------.-...........(((((((..((((------(.....((((((((((((((((((....)-....))))))))))).......)))))).....)))))..))))))).. ( -30.91) >DroSec_CAF1 9456 101 + 1 ---------U-UUUCAGCUGAAAAUGUCGUUGGUU------GCCAGUUAAAGUUGGUGACGAGGCGUG---AGGUCUUUGUCACUAGUGUUUUGCUUUGUGUUUCGGCUUUUGGCAUUAG ---------.-...........(((((((..((((------(.....((((((((((((((((((...---.)..))))))))))).......)))))).....)))))..))))))).. ( -29.21) >DroSim_CAF1 18283 101 + 1 ---------U-UUUCAGCUAAAAAUGUCGUUGGUU------GCCAGUUAAAGUUGGUGACGAGGCGUG---AGGUCUUUGUCACUAGUGUUUUGCUUUGUGUUUCGGCUUUUGGCAUUAG ---------.-.....(((((((..(((....((.------.((((......))))..))((((((..---((((......((....))....))))..))))))))))))))))..... ( -29.60) >DroEre_CAF1 15675 99 + 1 AUUUUCCGAUUUUGCUGCCGAAUAUGUCGCUUGUU------GCCAGUUAAAGUUGGCGCCGAG--------------UUGCCACUACAGUUGUGCUUUGU-UUUCAGCUUUUGGCAUUAG ...............(((((((...((.(((((.(------(((((......)))))).))))--------------).))......(((((........-...)))))))))))).... ( -24.00) >DroYak_CAF1 16233 120 + 1 AUUUCCCGAUUUUGCUGCUGAAAAUGCCGCUAGUUGGUUAUGCCGAUUAAAGUUGGUGACGAGGAGUGAGCAUGUUUUUGUCACUACUGUUUUGCUUUGUGUUUCGGCUUUUGGCAUUAG .....((((((..((.((.......)).)).))))))....(((((.((((((((((((((((((.........)))))))))))).......))))))....)))))............ ( -31.51) >consensus _________U_UUUCAGCUGAAAAUGUCGUUGGUU______GCCAGUUAAAGUUGGUGACGAGGCGUG___AGGUCUUUGUCACUACUGUUUUGCUUUGUGUUUCGGCUUUUGGCAUUAG ................((((((...(((((((((.......))))).(((((((((((((((((...........))))))))))).......)))))).....)))).))))))..... (-21.20 = -21.61 + 0.41)


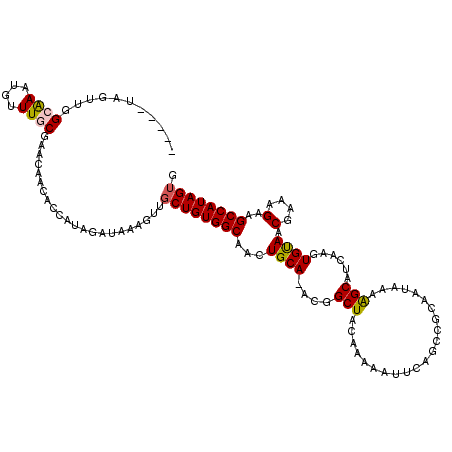
| Location | 8,466,780 – 8,466,899 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 92.72 |
| Mean single sequence MFE | -29.89 |
| Consensus MFE | -22.75 |
| Energy contribution | -22.12 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8466780 119 + 22224390 UCACUACUGUUUUGCUUUGUGUUUCGGCUUUUGGCAUUAGCACUAUGGCUUCUUUCGUUACACUUGAUGCUUUUAUUUCGGCUGCAUUUUUGUAGCCAUUGCAGUUGCCACAGCAACUU ...........(((((..(((((...((.....))...)))))..((((..((..(((((....)))))..........(((((((....))))))).....))..)))).)))))... ( -29.30) >DroSec_CAF1 9517 119 + 1 UCACUAGUGUUUUGCUUUGUGUUUCGGCUUUUGGCAUUAGCACUAUGGCUUCUUUCGUUGCACUCGAUGCUUUUAUUGCGGCUGAAUUUUUGUAGCCGUUGCAGUUGCCACAGCAACUU ...(.(((((...(((..(((((...((.....))...)))))...)))..........))))).).(((.......(((((((........))))))).)))(((((....))))).. ( -30.62) >DroSim_CAF1 18344 119 + 1 UCACUAGUGUUUUGCUUUGUGUUUCGGCUUUUGGCAUUAGCACUAUGGCUUCUUUCGUUACACUCGAUGCUUUUAUUGCGGCUGAAUUUUUGUAGCCGUUGCAGUUGCCACAGCAACUU ...(.(((((...(((..(((((...((.....))...)))))...)))..........))))).).(((.......(((((((........))))))).)))(((((....))))).. ( -30.32) >DroEre_CAF1 15735 118 + 1 CCACUACAGUUGUGCUUUGU-UUUCAGCUUUUGGCAUUAGCACUAUGGCUUCUUUCGUUACACUUGAUGCUUUCAUUCUGGCGGAAUUUUGGUAGCUGUUGCAGUUGCCACAGCAACUU ......((...(((((.(((-(..........))))..)))))..))(((..((((((((....(((.....)))...))))))))...(((((((((...))))))))).)))..... ( -29.30) >consensus UCACUACUGUUUUGCUUUGUGUUUCGGCUUUUGGCAUUAGCACUAUGGCUUCUUUCGUUACACUCGAUGCUUUUAUUGCGGCUGAAUUUUUGUAGCCGUUGCAGUUGCCACAGCAACUU ..........................(((..(((((...(((..((((((.(..(((.......))).(((........))).........).)))))))))...))))).)))..... (-22.75 = -22.12 + -0.62)


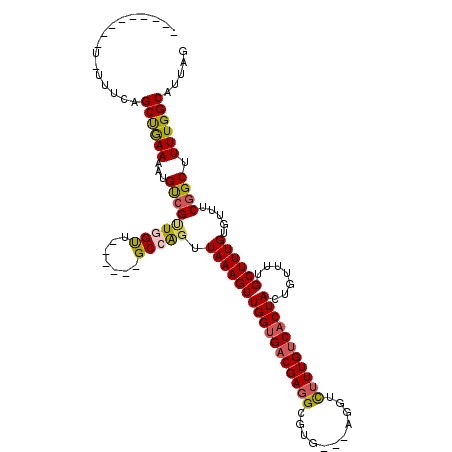
| Location | 8,466,780 – 8,466,899 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 92.72 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -20.66 |
| Energy contribution | -20.72 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8466780 119 - 22224390 AAGUUGCUGUGGCAACUGCAAUGGCUACAAAAAUGCAGCCGAAAUAAAAGCAUCAAGUGUAACGAAAGAAGCCAUAGUGCUAAUGCCAAAAGCCGAAACACAAAGCAAAACAGUAGUGA ...(((((((((((...((((((((((((...((((.............))))....)))).(....)..)))))..)))...))))....((...........))...)))))))... ( -28.82) >DroSec_CAF1 9517 119 - 1 AAGUUGCUGUGGCAACUGCAACGGCUACAAAAAUUCAGCCGCAAUAAAAGCAUCGAGUGCAACGAAAGAAGCCAUAGUGCUAAUGCCAAAAGCCGAAACACAAAGCAAAACACUAGUGA ...((((((((((...((((..((((..........))))((.......))......)))).(....)..))))).(((.....((.....)).....)))..)))))........... ( -26.00) >DroSim_CAF1 18344 119 - 1 AAGUUGCUGUGGCAACUGCAACGGCUACAAAAAUUCAGCCGCAAUAAAAGCAUCGAGUGUAACGAAAGAAGCCAUAGUGCUAAUGCCAAAAGCCGAAACACAAAGCAAAACACUAGUGA ...((((((((((...((((..((((..........))))((.......))......)))).(....)..))))).(((.....((.....)).....)))..)))))........... ( -24.00) >DroEre_CAF1 15735 118 - 1 AAGUUGCUGUGGCAACUGCAACAGCUACCAAAAUUCCGCCAGAAUGAAAGCAUCAAGUGUAACGAAAGAAGCCAUAGUGCUAAUGCCAAAAGCUGAAA-ACAAAGCACAACUGUAGUGG ..((.(((((.((....)).))))).)).......(((((((......(((((...(.((..(....)..)))...)))))..........(((....-....)))....)))..)))) ( -25.60) >consensus AAGUUGCUGUGGCAACUGCAACGGCUACAAAAAUUCAGCCGCAAUAAAAGCAUCAAGUGUAACGAAAGAAGCCAUAGUGCUAAUGCCAAAAGCCGAAACACAAAGCAAAACACUAGUGA .....(((.(((((...(((..((((.......................((((...))))..(....).))))....)))...)))))..))).......................... (-20.66 = -20.72 + 0.06)



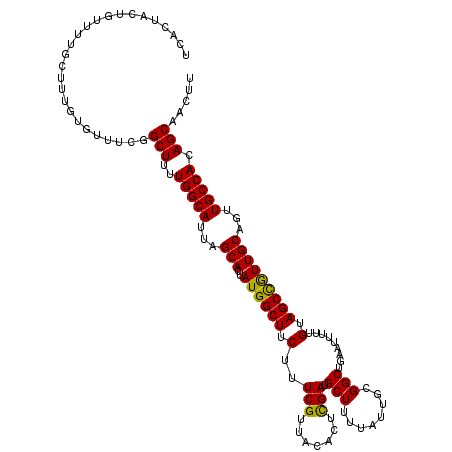
| Location | 8,466,820 – 8,466,934 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.93 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -16.69 |
| Energy contribution | -16.81 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8466820 114 - 22224390 -----UAGUUUGCAAACGUUUGCGAACAACACCAUAGAUAAAGUUGCUGUGGCAACUGCA-AUGGCUACAAAAAUGCAGCCGAAAUAAAAGCAUCAAGUGUAACGAAAGAAGCCAUAGUG -----..((((((((....))))))))..............((((((....))))))((.-(((((((((...((((.............))))....)))).(....)..))))).)). ( -30.22) >DroSec_CAF1 9557 108 - 1 -----UAGUUCGCAAAUGUUUGCGA------CCAUAGAUAAAGUUGCUGUGGCAACUGCA-ACGGCUACAAAAAUUCAGCCGCAAUAAAAGCAUCGAGUGCAACGAAAGAAGCCAUAGUG -----..(.((((((....))))))------.)............((((((((...((((-..((((..........))))((.......))......)))).(....)..)))))))). ( -30.90) >DroSim_CAF1 18384 114 - 1 -----UAGUUGGCAAAUGUUUGCGAACAACACCAUAGAUAAAGUUGCUGUGGCAACUGCA-ACGGCUACAAAAAUUCAGCCGCAAUAAAAGCAUCGAGUGUAACGAAAGAAGCCAUAGUG -----..(((.((((....)))).)))..................((((((((...((((-..((((..........))))((.......))......)))).(....)..)))))))). ( -29.30) >DroEre_CAF1 15774 111 - 1 --------UUUGCAAAUGUUUACCAAGAACACCAUUGAAGAAGUUGCUGUGGCAACUGCA-ACAGCUACCAAAAUUCCGCCAGAAUGAAAGCAUCAAGUGUAACGAAAGAAGCCAUAGUG --------........(((((.....)))))..............((((((((...((((-...(((......((((.....))))...)))......)))).(....)..)))))))). ( -20.00) >DroYak_CAF1 16353 119 - 1 CAAACUAUUUGGCGAAUGUUUACGAAUAAUGGCAUUGAAAAAGUUGCUGUGGCAACUGCAAACAGCUCCAAAAAUGCAGCCACAAUAAAGGCAGCAAGUGUAACGAAAGAAGCCAUAG-G ....(((..((((......(((((.....(((..(((....((((((....)))))).....)))..)))....(((.(((........))).)))..)))))(....)..)))))))-. ( -29.60) >consensus _____UAGUUGGCAAAUGUUUGCGAACAACACCAUAGAUAAAGUUGCUGUGGCAACUGCA_ACGGCUACAAAAAUUCAGCCGCAAUAAAAGCAUCAAGUGUAACGAAAGAAGCCAUAGUG ...........((((....))))......................((((((((...((((....(((......................)))......)))).(....)..)))))))). (-16.69 = -16.81 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:32 2006