
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,466,395 – 8,466,545 |
| Length | 150 |
| Max. P | 0.804699 |

| Location | 8,466,395 – 8,466,507 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -40.85 |
| Consensus MFE | -29.38 |
| Energy contribution | -30.75 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
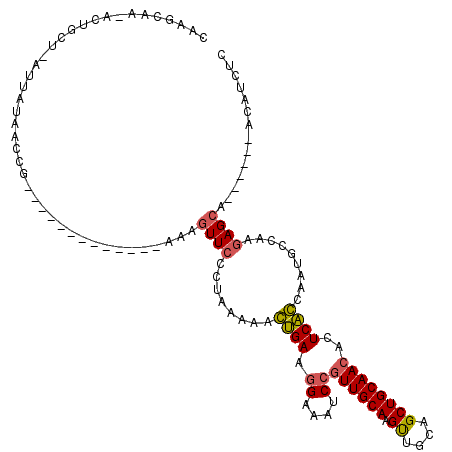
>X_DroMel_CAF1 8466395 112 + 22224390 GUCCGGAUUUGCAGGUUGCUAGUUGGGCAAAGCGGCUUGAGAUGU------UGCUCUUGGCAUUGCUGAGUGUUGCAGCUGCAACUUGCAAUGGAUUUCCUUCAGUUCUUAGAGAACU (((((...(((((((((((.(((((((((.(((.(((.(((....------..)))..)))...)))...)))).))))))))))))))))))))).......(((((.....))))) ( -41.50) >DroSec_CAF1 9176 112 + 1 GUGCGGAUUUGCAGGUUGCUAGUUGGGCAAAGCUGCUUGAGAUGU------UGCUCUUGGCAUUGGUGAGUGUUGCAGCUGCAACUUGCAACGGAUUUCCAUCACCUUUUAGGGAACU .....((((((((((((((.(((((((((..((((((.(((....------..)))..)))...)))...)))).)))))))))))))))).((....))))).(((....))).... ( -36.00) >DroSim_CAF1 17981 112 + 1 GUCCGGAUUUGCAGGUUGCUAGUUGGGCAAAGCUGCUUGAGAUGU------UGCUCUUGGCAUUGGUGAGUGUUGCAGCUGCAACUUGCAACGGAUUUCCAUCACUUUUUAGGGAACU (((((...(((((((((((.(((((((((..((((((.(((....------..)))..)))...)))...)))).)))))))))))))))))))))((((...........))))... ( -38.40) >DroYak_CAF1 15942 118 + 1 GGCCGGAUUGGCAGGUGGCAAGUUGGGCAAAGCUGCUUGAGAUGUGCAUGUGCCUCUUGGCGUUGCCGAGUGUUGCAGCUGCAGCUUGCAACGGAUUUCGUUCGGUGUUUGGCCAACU ((((((((.((((.((((((..(..((((....))))..)..))).))).)))).((((((...))))))(((((((((....)).))))))).............)))))))).... ( -47.50) >consensus GUCCGGAUUUGCAGGUUGCUAGUUGGGCAAAGCUGCUUGAGAUGU______UGCUCUUGGCAUUGCUGAGUGUUGCAGCUGCAACUUGCAACGGAUUUCCAUCACUUUUUAGGGAACU (((((....((((((((((.(((((((((.(((.(((.(((............)))..)))...)))...)))).))))))))))))))).)))))...................... (-29.38 = -30.75 + 1.38)


| Location | 8,466,395 – 8,466,507 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -29.21 |
| Consensus MFE | -18.54 |
| Energy contribution | -19.22 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.605155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8466395 112 - 22224390 AGUUCUCUAAGAACUGAAGGAAAUCCAUUGCAAGUUGCAGCUGCAACACUCAGCAAUGCCAAGAGCA------ACAUCUCAAGCCGCUUUGCCCAACUAGCAACCUGCAAAUCCGGAC (((((.....)))))...((....)).(((((.(((((.((((.......))))..((...(((((.------............)))))...))....))))).)))))........ ( -26.82) >DroSec_CAF1 9176 112 - 1 AGUUCCCUAAAAGGUGAUGGAAAUCCGUUGCAAGUUGCAGCUGCAACACUCACCAAUGCCAAGAGCA------ACAUCUCAAGCAGCUUUGCCCAACUAGCAACCUGCAAAUCCGCAC ............(((((.((....))((((((.((....))))))))..)))))..(((...(((..------....)))..((((..((((.......)))).))))......))). ( -29.30) >DroSim_CAF1 17981 112 - 1 AGUUCCCUAAAAAGUGAUGGAAAUCCGUUGCAAGUUGCAGCUGCAACACUCACCAAUGCCAAGAGCA------ACAUCUCAAGCAGCUUUGCCCAACUAGCAACCUGCAAAUCCGGAC .((((........(..((((....))))..)..(((((....)))))...............)))).------.........((((..((((.......)))).)))).......... ( -26.90) >DroYak_CAF1 15942 118 - 1 AGUUGGCCAAACACCGAACGAAAUCCGUUGCAAGCUGCAGCUGCAACACUCGGCAACGCCAAGAGGCACAUGCACAUCUCAAGCAGCUUUGCCCAACUUGCCACCUGCCAAUCCGGCC ....((((..................((((((.((....))))))))....((((..(.((((.((((..(((.........)))....))))...)))).)...)))).....)))) ( -33.80) >consensus AGUUCCCUAAAAACUGAAGGAAAUCCGUUGCAAGUUGCAGCUGCAACACUCACCAAUGCCAAGAGCA______ACAUCUCAAGCAGCUUUGCCCAACUAGCAACCUGCAAAUCCGGAC .............(((..((....)).(((((.(((((((((((......((....))....(((............)))..))))))...........))))).)))))...))).. (-18.54 = -19.22 + 0.69)



| Location | 8,466,433 – 8,466,545 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.56 |
| Mean single sequence MFE | -27.18 |
| Consensus MFE | -12.84 |
| Energy contribution | -12.78 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8466433 112 - 22224390 CAAGCAA-ACUGAU-AUUAUAACAGUAUUGUAUUGCGCAAAGUUCUCUAAGAACUGAAGGAAAUCCAUUGCAAGUUGCAGCUGCAACACUCAGCAAUGCCAAGAGCA------ACAUCUC ...((..-((((..-.......))))...((((((((((((((((.....)))))...((....)).))))..(((((....))))).....))))))).....)).------....... ( -30.30) >DroSec_CAF1 9214 99 - 1 CAAGCAA-ACUGCU-AUUAUAACGG-------------AAAGUUCCCUAAAAGGUGAUGGAAAUCCGUUGCAAGUUGCAGCUGCAACACUCACCAAUGCCAAGAGCA------ACAUCUC .......-..((((-........((-------------(....)))......(((((.((....))((((((.((....))))))))..))))).........))))------....... ( -23.00) >DroSim_CAF1 18019 99 - 1 CAAGCAA-ACUGCU-AUUAUAACCG-------------AAAGUUCCCUAAAAAGUGAUGGAAAUCCGUUGCAAGUUGCAGCUGCAACACUCACCAAUGCCAAGAGCA------ACAUCUC ..(((..-...)))-..........-------------...((((........(..((((....))))..)..(((((....)))))...............)))).------....... ( -21.10) >DroYak_CAF1 15980 107 - 1 CUAUUAAUGCUGCAUUGUAUGGCCG-------------AAAGUUGGCCAAACACCGAACGAAAUCCGUUGCAAGCUGCAGCUGCAACACUCGGCAACGCCAAGAGGCACAUGCACAUCUC ......(((.((((((((.((((((-------------(...))))))).)))((((..(.....)((((((.((....))))))))..))))....(((....)))..))))))))... ( -34.30) >consensus CAAGCAA_ACUGCU_AUUAUAACCG_____________AAAGUUCCCUAAAAACUGAAGGAAAUCCGUUGCAAGUUGCAGCUGCAACACUCACCAAUGCCAAGAGCA______ACAUCUC .........................................((((........((((.((....))((((((.((....))))))))..)))).........)))).............. (-12.84 = -12.78 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:28 2006