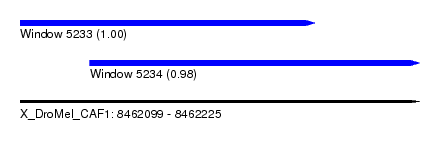
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,462,099 – 8,462,225 |
| Length | 126 |
| Max. P | 0.998904 |

| Location | 8,462,099 – 8,462,192 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.22 |
| Mean single sequence MFE | -25.93 |
| Consensus MFE | -22.00 |
| Energy contribution | -22.25 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.28 |
| SVM RNA-class probability | 0.998904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
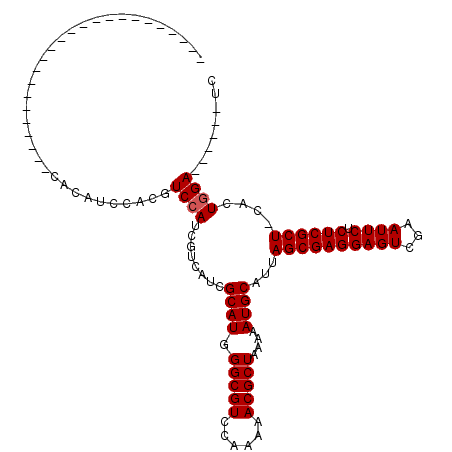
>X_DroMel_CAF1 8462099 93 + 22224390 CA------------------CAUCCACAUCCACGUCCAUCGUCAUCGCAUGGGCGUCCAAAAACGCUAAAAAUGCAUUAGCGAGGAGUCGAAUUCUCUCGCU-CACUGGA------UC ..------------------.......((((((((((((((....)).)))))))).......((((((.......))))))(((((.(((......)))))-).)))))------). ( -27.60) >DroSec_CAF1 5118 92 + 1 ------------------------CACAUCCACGUCCAUCGUCAUCGCAUAGGCGUCCAA-AACGCUAA-AAUGCAUUAGCGAGGAGUCGAAUUCUCUCGCUCCACUGGAAUCCCAUC ------------------------....((((.((...(((..(..((((.(((((....-.)))))..-.))))..)..)))((((.(((......)))))))))))))........ ( -25.70) >DroSim_CAF1 13871 87 + 1 ------------------------CACAUCCACGUCCAUCGUCAUCGCAUGGGCGUCCAAAAACGCUAAAAAUGCAUUAGCGAGGAGUCGAAUUCUCUCGCU-CACUGGA------UC ------------------------...((((((((((((((....)).)))))))).......((((((.......))))))(((((.(((......)))))-).)))))------). ( -27.60) >DroYak_CAF1 10511 111 + 1 CACAUCUCCACAUCUCCACAUCCACAUCUCCACAUCCAUCGUCAUCGCAUGGGCGUCCAAAAACGCUAAAAAUGCAUUAGCGAGGAGUCGAAUUCUCUCGCU-CACUCGA------UC .....................................((((.....((((.(((((......)))))....))))...((((((((((...)))).))))))-....)))------). ( -22.80) >consensus ________________________CACAUCCACGUCCAUCGUCAUCGCAUGGGCGUCCAAAAACGCUAAAAAUGCAUUAGCGAGGAGUCGAAUUCUCUCGCU_CACUGGA______UC ..................................((((........((((.(((((......)))))....))))...((((((((((...)))).))))))....))))........ (-22.00 = -22.25 + 0.25)

| Location | 8,462,121 – 8,462,225 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.93 |
| Mean single sequence MFE | -26.33 |
| Consensus MFE | -20.27 |
| Energy contribution | -20.27 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
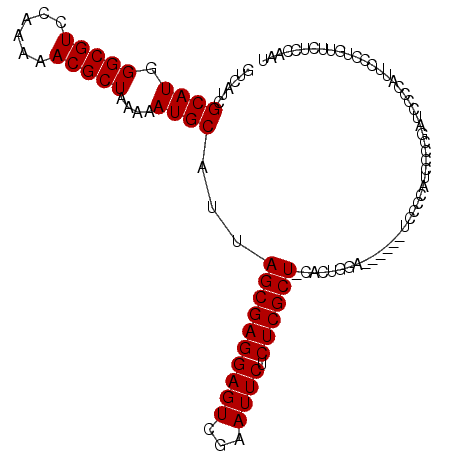
>X_DroMel_CAF1 8462121 104 + 22224390 GUCAUCGCAUGGGCGUCCAAAAACGCUAAAAAUGCAUUAGCGAGGAGUCGAAUUCUCUCGCU-CACUGGA------UCCCCAUCCCCGAUCCCCAUUCCCUAUUCUCCAAU ...((((.(((((.(((((....((((((.......))))))..(((.(((......)))))-)..))))------).)))))...))))..................... ( -27.90) >DroSec_CAF1 5134 109 + 1 GUCAUCGCAUAGGCGUCCAA-AACGCUAA-AAUGCAUUAGCGAGGAGUCGAAUUCUCUCGCUCCACUGGAAUCCCAUCACCAUCCCCGAUCCCAAUUCCCUGGUCUCCAAU ...........((..((((.-..((((((-......)))))).((((.(((......)))))))..))))...))............((..(((......)))..)).... ( -26.10) >DroSim_CAF1 13887 104 + 1 GUCAUCGCAUGGGCGUCCAAAAACGCUAAAAAUGCAUUAGCGAGGAGUCGAAUUCUCUCGCU-CACUGGA------UCCCCAUCUCCGAUCCCCAUUCCCUGUUCUCCAAU ...((((.(((((.(((((....((((((.......))))))..(((.(((......)))))-)..))))------).)))))...))))..................... ( -27.90) >DroYak_CAF1 10551 84 + 1 GUCAUCGCAUGGGCGUCCAAAAACGCUAAAAAUGCAUUAGCGAGGAGUCGAAUUCUCUCGCU-CACUCGA------UCCCCGCUCCCC--------------------GCU ......((((.(((((......)))))....)))).....(((((((.(((......)))))-).)))).------.....((.....--------------------)). ( -23.40) >consensus GUCAUCGCAUGGGCGUCCAAAAACGCUAAAAAUGCAUUAGCGAGGAGUCGAAUUCUCUCGCU_CACUGGA______UCCCCAUCCCCGAUCCCCAUUCCCUGUUCUCCAAU ......((((.(((((......)))))....))))...((((((((((...)))).))))))................................................. (-20.27 = -20.27 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:23 2006