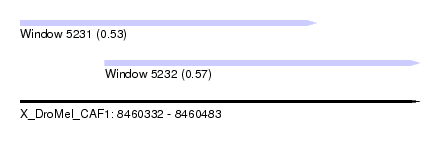
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,460,332 – 8,460,483 |
| Length | 151 |
| Max. P | 0.574698 |

| Location | 8,460,332 – 8,460,444 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 83.84 |
| Mean single sequence MFE | -37.42 |
| Consensus MFE | -25.85 |
| Energy contribution | -26.38 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8460332 112 + 22224390 GCUAGUUUCUUCAGACG-CGCACAAAGCUCGUCGCUCGGCACUCGUCACUCGGCACUCGGCACUCGAGCGUUCUUCGUUCGAGUGGCAGCUCUGGCAACUAAAUGAGCAAUUC ((((((.......((((-.((.....)).))))....(((....)))))).)))....(.((((((((((.....)))))))))).).((((.(....).....))))..... ( -39.70) >DroSec_CAF1 3403 98 + 1 GCUAGUUUCUUCAGACG-CGCACAAAGCUCGUCGCUCGGCACU--------------CGGCACUCGAGCGUUCUUCGUUCGAGUGGCAGCUCUGGCAACUAAAUGAGCAAUUC (((.((((.....((((-.((.....)).))))((.(((..((--------------.(.((((((((((.....)))))))))).)))..)))))....)))).)))..... ( -37.00) >DroSim_CAF1 12150 105 + 1 GCUAGUUUCUUCAGACG-CGCACAAAGCUCGUCGCUCGGCACU-------CGGGACUCGGCACUCGAGCGUUCUUCGUUCGAGUGGCAGCUCUGGCAACUAAAUGAGCAAUUC .............((((-.((.....)).))))(((((....(-------(((..((.(.((((((((((.....)))))))))).)))..))))........)))))..... ( -38.50) >DroEre_CAF1 9717 106 + 1 GCUGGUUUCUUCAGACGGCGCACAAAGCUCAUCGCUCGGCACUU-------GGCACUCGGCACUCGAGCGUUCUUCGUUCGAGUGGCAGCUCUGGCAACUAAAUGAGCAAUUC ((((...........)))).......((((((......((.(..-------(((....(.((((((((((.....)))))))))).).)))..)))......))))))..... ( -36.40) >DroYak_CAF1 8736 106 + 1 GCUGGUUUCUUCAGA-------CAAAGCUCGUCGCACGGCACUCGGUACUCGGUACUCGGCACUCGAGCGUUCUUCGUUCGAGUGGCAGCUCUGGCAGCUAAAUGAGCAAUUC .((((.....)))).-------....((((((.((...((..(((((((...)))).)))((((((((((.....)))))))))))).((....)).))...))))))..... ( -36.10) >DroAna_CAF1 17079 96 + 1 GCUUGGUUCUUUGGA------ACAAAGCUCGUUGCUCGUCGCUC-------GUUGUUUGGCACUCGAGCGUCCU----UCGAGUGGCAGCUCUGGAAUCUAAAUGAGCAAUUC .....((((((((((------.((.((((.((..((((.(((((-------(.((.....))..))))))....----.))))..)))))).))...)))))).))))..... ( -36.80) >consensus GCUAGUUUCUUCAGACG_CGCACAAAGCUCGUCGCUCGGCACUC_______GGCACUCGGCACUCGAGCGUUCUUCGUUCGAGUGGCAGCUCUGGCAACUAAAUGAGCAAUUC ..........................((((((..........................(.((((((((((.....)))))))))).).((....))......))))))..... (-25.85 = -26.38 + 0.53)



| Location | 8,460,364 – 8,460,483 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.51 |
| Mean single sequence MFE | -43.27 |
| Consensus MFE | -35.01 |
| Energy contribution | -35.77 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8460364 119 + 22224390 GCUCGGCACUCGUCACUCGGCACUCGGCACUCGAGCGUUCUUCGUUCGAGUGGCAGCUCUGGCAACUAAAUGAGCAAUUCAUGAGUUUGUUGGCAACCCUACUUGAGGAUGGUCGCAGU- (((((((....((((..(((.(((((.((((((((((.....)))))))))).).((((.(....).....)))).......))))))).))))...(((.....)))...)))).)))- ( -42.80) >DroSec_CAF1 3435 105 + 1 GCUCGGCACU--------------CGGCACUCGAGCGUUCUUCGUUCGAGUGGCAGCUCUGGCAACUAAAUGAGCAAUUCAUGAGUUUGUUGGCAACCCUACUUGAGGGUGGUCGCAGU- .....(((((--------------((.((((((((((.....)))))))))).).((((.(....).....)))).......)))).....(((.(((((.....))))).)))))...- ( -42.30) >DroSim_CAF1 12182 112 + 1 GCUCGGCACU-------CGGGACUCGGCACUCGAGCGUUCUUCGUUCGAGUGGCAGCUCUGGCAACUAAAUGAGCAAUUCAUGAGUUUGUUGGCAACCCUACUUGAGGGUGGUCGCAGU- .....((...-------..(((((((.((((((((((.....)))))))))).).((((.(....).....)))).......))))))...(((.(((((.....))))).)))))...- ( -44.80) >DroEre_CAF1 9750 112 + 1 GCUCGGCACUU-------GGCACUCGGCACUCGAGCGUUCUUCGUUCGAGUGGCAGCUCUGGCAACUAAAUGAGCAAUUCAUGAGUUUGUUGGCAACCCUACUUGAGGGUGGUCGCGGC- (((((((..(.-------.(((((((.((((((((((.....)))))))))).).((((.(....).....)))).......))))..))..)..(((((.....))))).)))).)))- ( -44.70) >DroYak_CAF1 8762 119 + 1 GCACGGCACUCGGUACUCGGUACUCGGCACUCGAGCGUUCUUCGUUCGAGUGGCAGCUCUGGCAGCUAAAUGAGCAAUUCAUGAGUUUGUUGGCAACCCUACUUGAGGGUGGUCGCAGC- ((((((.(((((((((...))))).(.((((((((((.....)))))))))).).((((............)))).......)))))))).(((.(((((.....))))).)))))...- ( -44.10) >DroAna_CAF1 17106 109 + 1 GCUCGUCGCUC-------GUUGUUUGGCACUCGAGCGUCCU----UCGAGUGGCAGCUCUGGAAUCUAAAUGAGCAAUUCAUGGGUUUGUUGGCAACCCUACUUGAGGGCUGCCCCUCUC (((((.(((((-------(.((.....))..))))))....----.)))))(((((((((.((......(((((...)))))(((((.(....))))))...)).)))))))))...... ( -40.90) >consensus GCUCGGCACUC_______GGCACUCGGCACUCGAGCGUUCUUCGUUCGAGUGGCAGCUCUGGCAACUAAAUGAGCAAUUCAUGAGUUUGUUGGCAACCCUACUUGAGGGUGGUCGCAGU_ (((((....................(.((((((((((.....)))))))))).).((((.(....).....))))......))))).....(((.(((((.....))))).)))...... (-35.01 = -35.77 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:21 2006