
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,459,198 – 8,459,288 |
| Length | 90 |
| Max. P | 0.962547 |

| Location | 8,459,198 – 8,459,288 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.79 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -20.91 |
| Energy contribution | -21.05 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8459198 90 + 22224390 G--GCUAAUUAAAUCAAGUGCUCGGCGGGUUUUCCUCGA------UUC---AACUGCGUCAUUGUUUGGACAAUGGCUGAGCAAUUAGCCACCGAUUC-------------------CAU (--((((((.........(((((((((((((......))------)))---........((((((....)))))))))))))))))))))........-------------------... ( -28.10) >DroPse_CAF1 7478 85 + 1 UGCGCUAAUUAAAUCGAGUGCUCAGCGGGUUUUCCUUUU------UUC---GACUGCGUCAUUGUUUGGACAAUGGCUGAGCAAUUAGCCGCCG-------------------------- .((((((((.........((((((((((...........------...---..))))((((((((....))))))))))))))))))).)))..-------------------------- ( -26.69) >DroEre_CAF1 8519 90 + 1 G--GCUAAUUAAAUCAAGUGCUCGGCGGGUUUUCCUCGA------UUC---AACUGCGUCAUUGUUUGGACAAUGGCUGAGCAAUUAGCCACCGAUUC-------------------CCC (--((((((.........(((((((((((((......))------)))---........((((((....)))))))))))))))))))))........-------------------... ( -28.10) >DroYak_CAF1 7316 115 + 1 G--GCUAAUUAAAUCAAGUGCUCGUCGGGCUUUCCUCGGCGGCUUUUC---AACUGCGUCAUUGUUUGGACAAUGGCUGAGCAAUUAGCCACCGAUUCCGAUUCCGAUUCCCUAUUUCCC (--(((((((...(((...((.(((((((.....))))))))).....---......((((((((....)))))))))))..)))))))).............................. ( -32.80) >DroAna_CAF1 15548 103 + 1 AGCGCUAAUUAAAUCAAGUGCCUGGCGGGUUUUCCUUUUCCUCAUUUUUGUGCCGUCGUCAUUGUUUGGACAAUGGCUGAGCAAUUAGCCAUCACUGAC--C---------------GCC .(((((..........)))))..(((((...................((((.(((.((....))..)))))))(((((((....))))))).......)--)---------------))) ( -28.30) >DroPer_CAF1 8739 85 + 1 UGCGCUAAUUAAAUCGAGUGCUCAGCGGGUUUUCCUUUU------UUC---GACUGCGUCAUUGUUUGGACAAUGGCUGAGAAAUUAGCCGCCG-------------------------- .(((((((((..........((((((((...........------...---..))))((((((((....)))))))))))).)))))).)))..-------------------------- ( -22.69) >consensus G__GCUAAUUAAAUCAAGUGCUCGGCGGGUUUUCCUCGU______UUC___AACUGCGUCAUUGUUUGGACAAUGGCUGAGCAAUUAGCCACCGAUUC___________________CCC ...((((((.........(((((((((((....))).......................((((((....))))))))))))))))))))............................... (-20.91 = -21.05 + 0.14)


| Location | 8,459,198 – 8,459,288 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.79 |
| Mean single sequence MFE | -27.05 |
| Consensus MFE | -16.37 |
| Energy contribution | -16.73 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
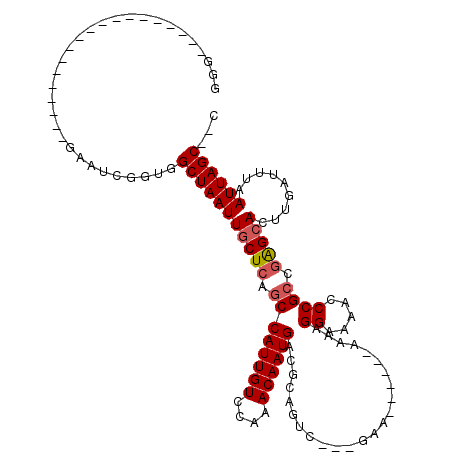
>X_DroMel_CAF1 8459198 90 - 22224390 AUG-------------------GAAUCGGUGGCUAAUUGCUCAGCCAUUGUCCAAACAAUGACGCAGUU---GAA------UCGAGGAAAACCCGCCGAGCACUUGAUUUAAUUAGC--C .((-------------------((..((((((((........)))))))))))).........((((((---(((------(((((................)))))))))))).))--. ( -26.59) >DroPse_CAF1 7478 85 - 1 --------------------------CGGCGGCUAAUUGCUCAGCCAUUGUCCAAACAAUGACGCAGUC---GAA------AAAAGGAAAACCCGCUGAGCACUCGAUUUAAUUAGCGCA --------------------------..(((.(((((((((((((((((((....))))))........---...------....((.....)))))))))).........)))))))). ( -26.30) >DroEre_CAF1 8519 90 - 1 GGG-------------------GAAUCGGUGGCUAAUUGCUCAGCCAUUGUCCAAACAAUGACGCAGUU---GAA------UCGAGGAAAACCCGCCGAGCACUUGAUUUAAUUAGC--C ..(-------------------((..((((((((........)))))))))))..........((((((---(((------(((((................)))))))))))).))--. ( -25.59) >DroYak_CAF1 7316 115 - 1 GGGAAAUAGGGAAUCGGAAUCGGAAUCGGUGGCUAAUUGCUCAGCCAUUGUCCAAACAAUGACGCAGUU---GAAAAGCCGCCGAGGAAAGCCCGACGAGCACUUGAUUUAAUUAGC--C .((.......(((((((..(((...(((((((((((((((...(.((((((....)))))).)))))))---....)))))))))((.....))..)))....)))))))......)--) ( -33.52) >DroAna_CAF1 15548 103 - 1 GGC---------------G--GUCAGUGAUGGCUAAUUGCUCAGCCAUUGUCCAAACAAUGACGACGGCACAAAAAUGAGGAAAAGGAAAACCCGCCAGGCACUUGAUUUAAUUAGCGCU (((---------------(--(((((((.((((...((.(((((((.(((((........))))).))).......)))).))..((.....))))))..))).))))........)))) ( -28.41) >DroPer_CAF1 8739 85 - 1 --------------------------CGGCGGCUAAUUUCUCAGCCAUUGUCCAAACAAUGACGCAGUC---GAA------AAAAGGAAAACCCGCUGAGCACUCGAUUUAAUUAGCGCA --------------------------..(((.((((((.((((((((((((....))))))........---...------....((.....))))))))..........))))))))). ( -21.90) >consensus GGG___________________GAAUCGGUGGCUAAUUGCUCAGCCAUUGUCCAAACAAUGACGCAGUC___GAA______AAAAGGAAAACCCGCCGAGCACUUGAUUUAAUUAGC__C ...............................(((((((((((.((((((((....))))))........................((.....)))).))))).........))))))... (-16.37 = -16.73 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:19 2006