
| Sequence ID | X_DroMel_CAF1 |
|---|---|
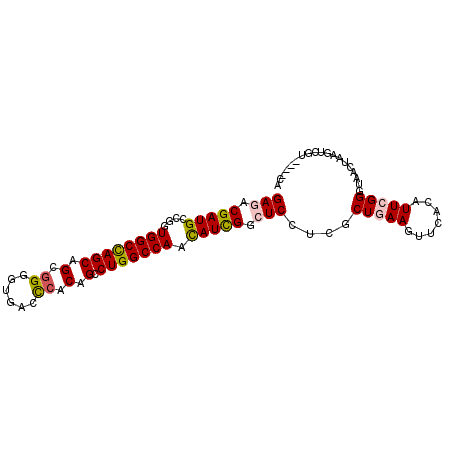
| Location | 8,442,873 – 8,442,969 |
| Length | 96 |
| Max. P | 0.649441 |

| Location | 8,442,873 – 8,442,969 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 81.63 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -21.96 |
| Energy contribution | -22.02 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8442873 96 + 22224390 GAAACGAUGCCGGUGGCCAGCAGCGGUGUGACCAACAGCCUGGCCAACAUCGGAUCCUCGCUGAAGUUCACGUUCGGGUAACUCCGGCGUUCCACA .....((((....(((((((..((.((.......)).))))))))).))))(((....(((((.((((.((......)))))).))))).)))... ( -33.30) >DroVir_CAF1 5191 90 + 1 GAGACGAUGCCAGUGGCCAGCAGCGGGGUUACCCACAGUCUGGCCAACAUUGGCUCCUCGCUGAAGUUUACAUUCGGGUAACUAAGUCGU------ ..(((((.(((((((((((((.(.(((....))).).).)))))))...)))))))....(((((.......)))))........)))..------ ( -32.90) >DroGri_CAF1 62036 92 + 1 GAGACGAUGCCAGUGGCCAGCAGUGGGGUCACCCACAGUCUGGCCAAUAUUGGCUCCUCGCUCAAGUUCACUUUCGGGUAACUAAAUCGC----CA (((.(((.(((((((((((((.(((((....))))).).)))))))...)))))...)))))).((((.(((....))))))).......----.. ( -38.10) >DroMoj_CAF1 95348 90 + 1 GAGACGAUGCCCGUGGCCAGCAGCGGCGUCACCCACAGCCUGGCCAACAUUGGCUCCUCGCUCAAAUUCACAUUCGGGUAACUAAGUCGC------ ..(((..((((((((((((((.(.((......)).).).)))))))...(((((.....)).))).........)))))).....)))..------ ( -28.40) >DroAna_CAF1 4185 96 + 1 GAGACGAUGCCGGUGGCUAGCAGCGGAGUGACUCACAGCCUGGCCAAUAUCGGCUCCUCGCUGAAGUUCACGUUCGGGUAACUUCGGCCUUCCAAA (((..((.((((((((((((..((.(((...)))...)))))))))...))))))))))(((((((((.((......)))))))))))........ ( -35.80) >DroPer_CAF1 5227 91 + 1 GAGACGAUGCCGGUGGCCAGCAGUGGGGUGACCCACAGCCUGGCCAAUAUCGGAUCAUCGCUGAAGUUUACAUUCGGGUAAC-----CAUCCAUCG .((.(((((((((((((((((.(((((....))))).).)))))))...))))..)))))))(((.......)))((((...-----.)))).... ( -39.10) >consensus GAGACGAUGCCGGUGGCCAGCAGCGGGGUGACCCACAGCCUGGCCAACAUCGGCUCCUCGCUGAAGUUCACAUUCGGGUAACUAAGUCGU____CA (((.(((((....((((((((.(.((......)).).).))))))).))))).)))....(((((.......)))))................... (-21.96 = -22.02 + 0.06)


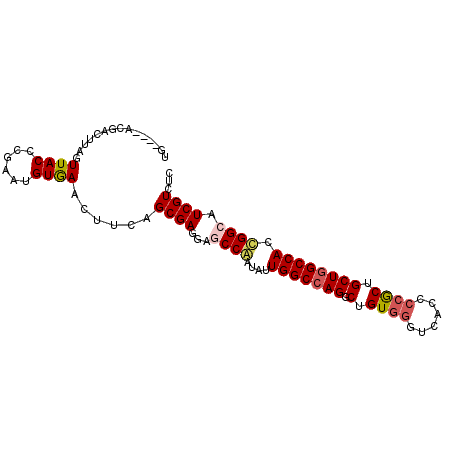
| Location | 8,442,873 – 8,442,969 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 81.63 |
| Mean single sequence MFE | -34.05 |
| Consensus MFE | -24.99 |
| Energy contribution | -25.22 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8442873 96 - 22224390 UGUGGAACGCCGGAGUUACCCGAACGUGAACUUCAGCGAGGAUCCGAUGUUGGCCAGGCUGUUGGUCACACCGCUGCUGGCCACCGGCAUCGUUUC .......(((.((((((((......))).))))).)))..((..(((((((((((((.(.((.((.....)))).))))))))...))))))..)) ( -34.40) >DroVir_CAF1 5191 90 - 1 ------ACGACUUAGUUACCCGAAUGUAAACUUCAGCGAGGAGCCAAUGUUGGCCAGACUGUGGGUAACCCCGCUGCUGGCCACUGGCAUCGUCUC ------..(((.......((((..((.......)).)).)).((((....(((((((...(((((....)))))..))))))).))))...))).. ( -32.70) >DroGri_CAF1 62036 92 - 1 UG----GCGAUUUAGUUACCCGAAAGUGAACUUGAGCGAGGAGCCAAUAUUGGCCAGACUGUGGGUGACCCCACUGCUGGCCACUGGCAUCGUCUC .(----(((((.......((((.(((....)))...)).)).((((....(((((((...(((((....)))))..))))))).)))))))))).. ( -40.90) >DroMoj_CAF1 95348 90 - 1 ------GCGACUUAGUUACCCGAAUGUGAAUUUGAGCGAGGAGCCAAUGUUGGCCAGGCUGUGGGUGACGCCGCUGCUGGCCACGGGCAUCGUCUC ------((.......((((......))))......))((.(((((...((.((((((.(.((((......)))).))))))))).))).)).)).. ( -31.02) >DroAna_CAF1 4185 96 - 1 UUUGGAAGGCCGAAGUUACCCGAACGUGAACUUCAGCGAGGAGCCGAUAUUGGCCAGGCUGUGAGUCACUCCGCUGCUAGCCACCGGCAUCGUCUC ........((.((((((((......))).))))).))((.((((((....((((..(((.(((........))).))).)))).)))).)).)).. ( -30.00) >DroPer_CAF1 5227 91 - 1 CGAUGGAUG-----GUUACCCGAAUGUAAACUUCAGCGAUGAUCCGAUAUUGGCCAGGCUGUGGGUCACCCCACUGCUGGCCACCGGCAUCGUCUC .(.((((..-----.((((......))))..)))).)(((((((((....(((((((.(.(((((....))))).)))))))).))).)))))).. ( -35.30) >consensus UG____ACGACUUAGUUACCCGAAUGUGAACUUCAGCGAGGAGCCAAUAUUGGCCAGGCUGUGGGUCACCCCGCUGCUGGCCACCGGCAUCGUCUC ...............((((......))))......((((...((((....(((((((.(.((((......)))).)))))))).)))).))))... (-24.99 = -25.22 + 0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:11 2006