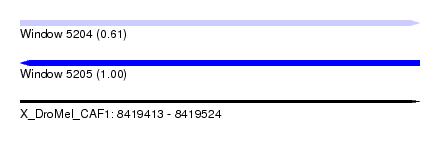
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,419,413 – 8,419,524 |
| Length | 111 |
| Max. P | 0.997874 |

| Location | 8,419,413 – 8,419,524 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.77 |
| Mean single sequence MFE | -42.12 |
| Consensus MFE | -24.42 |
| Energy contribution | -24.58 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8419413 111 + 22224390 AAGCUUUUGUUGUAGUGUAAC---------CGGAGCAUCAGCGCGUGAGCUUUUCGGUAUUCACGCUGCACUGACGGAUGGUCACACUAUGCGCGGCAUUUGGCAUUUUUGGCCGCGCAA ..........((((((((.((---------((((((....))((....))..))))))....)))))))).((((.....)))).....((((((((..............)))))))). ( -39.94) >DroSec_CAF1 4682 111 + 1 AAGCUUUUGUUGUAGUGUAAC---------CGAAGCAUCCGCGCGUGAGCUUUUUGGUAUUUACGCCUCACUGGCGGAUGGUCACACUGUGCGCGGCAUUUGGCAUUUUUGGCCGCGCAA ............((((((.((---------(.....((((((((....)).....(((......)))......))))))))).))))))((((((((..............)))))))). ( -40.74) >DroSim_CAF1 2323 110 + 1 AAGCUUUUGUUGUAGUGUAAC---------CGAA-CAUCCGCGCGUGAGCUUUUUGGUAUUUACGCCUCACUGGCGGAUGGUCACACUGUGCGCGGCAUUUGGCAUUUUUGGCCGCGCAA ............((((((.((---------(...-.((((((((....)).....(((......)))......))))))))).))))))((((((((..............)))))))). ( -41.14) >DroEre_CAF1 4997 111 + 1 AAGCUUUCGCUGUAGUGUGAC---------GGUAACAGCAGCGUGCGAGCUUUUUGGUAUUUACGCUGCACCGGCGAACGGUCACACUGCGCUCGGCUUUUGGUAUUUUUGGCCGCGCAA .(((....)))((((((((((---------.......((((((((.(.(((....))).).))))))))..((.....))))))))))))((.(((((............))))).)).. ( -45.80) >DroYak_CAF1 2326 120 + 1 AAGCUUUCGUUAUAAUGUGACCGUAGCGACCGUAACAGCAGCGUGCGAGCUUUUCAGUAUUUACGCUGCGCUGACGGACGGUCACACUGCACGCGACUUUUGGCAUUUUUGGCCGCGCAA ..((..((((..((.(((((((((.....((((....((((((((.(.(((....))).).))))))))....))))))))))))).))...)))).....(((.......))))).... ( -43.00) >consensus AAGCUUUUGUUGUAGUGUAAC_________CGAAACAUCAGCGCGUGAGCUUUUUGGUAUUUACGCUGCACUGGCGGAUGGUCACACUGUGCGCGGCAUUUGGCAUUUUUGGCCGCGCAA .(((....)))(((((((........................(((((((((....)))..))))))......(((.....))))))))))(((((((..............))))))).. (-24.42 = -24.58 + 0.16)


| Location | 8,419,413 – 8,419,524 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.77 |
| Mean single sequence MFE | -42.36 |
| Consensus MFE | -30.71 |
| Energy contribution | -29.07 |
| Covariance contribution | -1.64 |
| Combinations/Pair | 1.39 |
| Mean z-score | -3.73 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8419413 111 - 22224390 UUGCGCGGCCAAAAAUGCCAAAUGCCGCGCAUAGUGUGACCAUCCGUCAGUGCAGCGUGAAUACCGAAAAGCUCACGCGCUGAUGCUCCG---------GUUACACUACAACAAAAGCUU .((((((((..............))))))))((((((((((...(((((((...((((((............)))))))))))))....)---------)))))))))............ ( -44.44) >DroSec_CAF1 4682 111 - 1 UUGCGCGGCCAAAAAUGCCAAAUGCCGCGCACAGUGUGACCAUCCGCCAGUGAGGCGUAAAUACCAAAAAGCUCACGCGCGGAUGCUUCG---------GUUACACUACAACAAAAGCUU .((((((((..............)))))))).(((((((((((((((..(((((.(..............))))))..)))))).....)---------))))))))............. ( -41.78) >DroSim_CAF1 2323 110 - 1 UUGCGCGGCCAAAAAUGCCAAAUGCCGCGCACAGUGUGACCAUCCGCCAGUGAGGCGUAAAUACCAAAAAGCUCACGCGCGGAUG-UUCG---------GUUACACUACAACAAAAGCUU .((((((((..............)))))))).(((((((((((((((..(((((.(..............))))))..)))))).-...)---------))))))))............. ( -42.18) >DroEre_CAF1 4997 111 - 1 UUGCGCGGCCAAAAAUACCAAAAGCCGAGCGCAGUGUGACCGUUCGCCGGUGCAGCGUAAAUACCAAAAAGCUCGCACGCUGCUGUUACC---------GUCACACUACAGCGAAAGCUU ..(((((((..............)))..))))((((((((.((..((.((((((((..............))).)))).).))....)).---------))))))))..(((....))). ( -37.88) >DroYak_CAF1 2326 120 - 1 UUGCGCGGCCAAAAAUGCCAAAAGUCGCGUGCAGUGUGACCGUCCGUCAGCGCAGCGUAAAUACUGAAAAGCUCGCACGCUGCUGUUACGGUCGCUACGGUCACAUUAUAACGAAAGCUU ..(((((((..............)))))))..(((((((((((((((.((((((((((.....((....)).....))))))).))))))).....)))))))))))....(....)... ( -45.54) >consensus UUGCGCGGCCAAAAAUGCCAAAUGCCGCGCACAGUGUGACCAUCCGCCAGUGCAGCGUAAAUACCAAAAAGCUCACGCGCUGAUGCUACG_________GUUACACUACAACAAAAGCUU .((((((((..............)))))))).((((((((((((.((..(((.(((..............))))))..)).))))..............))))))))............. (-30.71 = -29.07 + -1.64)

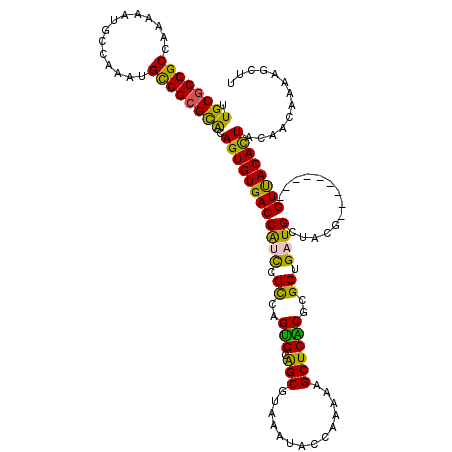

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:57 2006