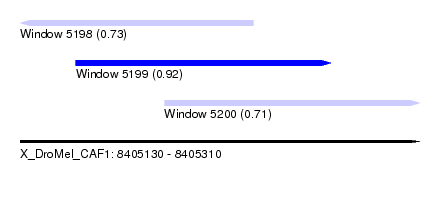
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,405,130 – 8,405,310 |
| Length | 180 |
| Max. P | 0.917516 |

| Location | 8,405,130 – 8,405,235 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -30.23 |
| Consensus MFE | -23.29 |
| Energy contribution | -24.60 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8405130 105 - 22224390 CCUGAAAUCAAGAGACUUUGCCUCGCCACCGAGAAUAGUCAUAUCGCAGCCAAUGCCAUGCUGCUAACAAAGAUAGAUGGUAUGUUUGUGACAUAC---------------AUAUCUUAU .............((((....((((....))))...)))).....(((((.........))))).....((((((....(((((((...)))))))---------------.)))))).. ( -25.30) >DroSim_CAF1 7450 101 - 1 CCUGAAAUCAAGAGGCUUUGCCUCGCCACCGAGCACAGUCAUAUCGCAGCCAAUGCCAUGCUGCUGACAAAGAUAGAUGGUAUGUUUGUGACA-------------------UAUCUUAU .........((((((((.(((.(((....)))))).))))...((((((...(((((((.(((((.....)).))))))))))..))))))..-------------------..)))).. ( -28.00) >DroEre_CAF1 7795 120 - 1 CCUGAAAUAAAAAGGCUUUGCCUCGCCACCGAGCAAAGUCAUAUCGCAGCCAAUGCCAUGAUGCUGACAAAGCUAGAUGGUAGGUUUGUAGCAUUUUGAUGUUUGGUCAUCAUAUCUAAC ..(((........((((((((.(((....)))))))))))........((((((..((.(((((((.((((.(((.....))).))))))))))).))..).)))))..)))........ ( -33.20) >DroYak_CAF1 7429 120 - 1 CCUGAAAUAAAGAGGCUUUGCCUCGCCACCGAGCAAAGUCAUAUCGCAGCGAAUGCCAUGAUGCUGACAAAGCUAGAUGGUAUGUUUGUGGCAUUUUGAUGUUUUGUCAUCACAUCUAAA ..........(((((((((((.(((....))))))))))).....((.(((((((((((...(((.....)))...)))))..)))))).))....(((((......)))))..)))... ( -34.40) >consensus CCUGAAAUAAAGAGGCUUUGCCUCGCCACCGAGCAAAGUCAUAUCGCAGCCAAUGCCAUGAUGCUGACAAAGAUAGAUGGUAUGUUUGUGACAUUU_______________AUAUCUAAU .............((((((((.(((....)))))))))))...((((((...(((((((...(((.....)))...)))))))..))))))............................. (-23.29 = -24.60 + 1.31)


| Location | 8,405,155 – 8,405,270 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.61 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -28.77 |
| Energy contribution | -29.40 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8405155 115 + 22224390 CCAUCUAUCUUUGUUAGCAGCAUGGCAUUGGCUGCGAUAUGACUAUUCUCGGUGGCGAGGCAAAGUCUCUUGAUUUCAGGCAUUGCUUCGUUAAUUUUUAGCAUUAAAG-----UGAAUA ...((...(((((...(((((.........)))))...(((.(((.......(((((((((((.((((.........)))).))))))))))).....)))))))))))-----.))... ( -29.50) >DroSim_CAF1 7471 114 + 1 CCAUCUAUCUUUGUCAGCAGCAUGGCAUUGGCUGCGAUAUGACUGUGCUCGGUGGCGAGGCAAAGCCUCUUGAUUUCAGGAGUUGUUUCGUUAAUUUUCAGCAUUAAAG-----UGAAU- ...((...(((((((((((((.........)))))....)))).(((((...(((((((((((...((((((....)))))))))))))))))......))))).))))-----.))..- ( -33.30) >DroEre_CAF1 7835 119 + 1 CCAUCUAGCUUUGUCAGCAUCAUGGCAUUGGCUGCGAUAUGACUUUGCUCGGUGGCGAGGCAAAGCCUUUUUAUUUCAGGCAUUGCUUCGUUCAUGUUGGACAUUAAAUUAAAGUGACU- (((((.(((...((((..(((..(((....)))..))).))))...))).))))).(((((((.((((.........)))).)))))))((((.....)))).................- ( -39.20) >DroYak_CAF1 7469 114 + 1 CCAUCUAGCUUUGUCAGCAUCAUGGCAUUCGCUGCGAUAUGACUUUGCUCGGUGGCGAGGCAAAGCCUCUUUAUUUCAGGUAUUGCUUCGCUUAUGUUGGACAUUAAAC-----UGAAU- .((((.(((...((((..(((..(((....)))..))).))))...))).))))(((((((((.((((.........)))).)))))))))....(((........)))-----.....- ( -37.60) >consensus CCAUCUAGCUUUGUCAGCAGCAUGGCAUUGGCUGCGAUAUGACUUUGCUCGGUGGCGAGGCAAAGCCUCUUGAUUUCAGGCAUUGCUUCGUUAAUGUUGAACAUUAAAG_____UGAAU_ .((((.(((...((((..(((..(((....)))..))).))))...))).))))(((((((((.((((.........)))).)))))))))............................. (-28.77 = -29.40 + 0.63)



| Location | 8,405,195 – 8,405,310 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.71 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -17.26 |
| Energy contribution | -17.82 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8405195 115 + 22224390 GACUAUUCUCGGUGGCGAGGCAAAGUCUCUUGAUUUCAGGCAUUGCUUCGUUAAUUUUUAGCAUUAAAG-----UGAAUACAUAUGUAUAUUUAGUUUAUGAAUUGAGGAAAGUCCCUAA ((((.((((((((((((((((((.((((.........)))).)))))))))).........(((.(((.-----((((((........)))))).)))))).)))))))).))))..... ( -32.60) >DroSim_CAF1 7511 107 + 1 GACUGUGCUCGGUGGCGAGGCAAAGCCUCUUGAUUUCAGGAGUUGUUUCGUUAAUUUUCAGCAUUAAAG-----UGAAU--------AUAUUGAGUUUACGAAUGGAGGAAAAUCCCUAA ....(((((...(((((((((((...((((((....)))))))))))))))))......)))))....(-----(((((--------.......))))))...(((.((.....))))). ( -28.40) >DroEre_CAF1 7875 112 + 1 GACUUUGCUCGGUGGCGAGGCAAAGCCUUUUUAUUUCAGGCAUUGCUUCGUUCAUGUUGGACAUUAAAUUAAAGUGACU--------AUAUUUAUUUUUUGAAUUGGGAAAAAUCCUUAG ((.(((.((((((.(((((((((.((((.........)))).)))))))((((.....))))...........)).)))--------..(((((.....))))).))).))).))..... ( -25.60) >DroYak_CAF1 7509 107 + 1 GACUUUGCUCGGUGGCGAGGCAAAGCCUCUUUAUUUCAGGUAUUGCUUCGCUUAUGUUGGACAUUAAAC-----UGAAU--------AUAUUUAAUUUUUGAGUAGGAAAAAAUCCUUAA .....(((((((.((((((((((.((((.........)))).))))))))))((((((((........)-----).)))--------)))........)))))))(((.....))).... ( -27.80) >consensus GACUUUGCUCGGUGGCGAGGCAAAGCCUCUUGAUUUCAGGCAUUGCUUCGUUAAUGUUGAACAUUAAAG_____UGAAU________AUAUUUAGUUUAUGAAUUGAGAAAAAUCCCUAA .....(((((((.((((((((((.((((.........)))).))))))))))....)))))))......................................................... (-17.26 = -17.82 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:52 2006