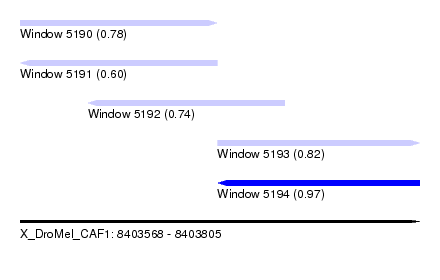
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,403,568 – 8,403,805 |
| Length | 237 |
| Max. P | 0.973080 |

| Location | 8,403,568 – 8,403,685 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -46.60 |
| Consensus MFE | -37.70 |
| Energy contribution | -38.45 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8403568 117 + 22224390 GUCCUCAUAGGAUAUCUCAUCGGUUAUCUUGACCAAGAGCUCCAGCAGUUCCCUGCUAUUCGCGGCCGUCAUUGGGGGAGCUCCAGUCGC---CGCAGCCGGCGGAUUUCCGAUGACACC ((((.....))))...(((((((..((((.(((...((((((((((((....)))))........(((....))).)))))))..)))((---((....))))))))..))))))).... ( -48.10) >DroSec_CAF1 2774 117 + 1 GUCCUCGUAGGAUAUCUCAUCGGUGAUCUUGACCAAGAGCUCCAGCAGUUCUCUGCUAUUAGCGGCUGUCAUUGGAGGAGCUCCAGUCGC---CGCAGCCGGCGGAUUUCCGAUGACACC ((((.....))))...(((((((.(((((.(.((....(((..(((((....)))))...)))((((((.(((((((...)))))))...---.)))))))))))))).))))))).... ( -47.10) >DroEre_CAF1 6221 117 + 1 GUCCUCGUAGGAUAUCUCAUCGGUGAUCUUGACCAAGAGCUCCAGCAGUUCCCUGCUGUUGCCGGCGGUCAUUUGGGGAGCUCCCGUCGC---CGCGGCCGGCUGAUUUCCGAGGGCAGC ((((((...(((.(((.....((((((..(((((..(.((..((((((....))))))..)))...)))))....(((....))))))))---)((.....)).))).)))))))))... ( -47.70) >DroYak_CAF1 5859 120 + 1 GUCCUCGUAGGAUAUCUCAUCGGUGAUUUUGACCAAUAGCUCCAGUAGUUCCCUGCUAUUGCCGGCCGUCAUUUGAGGAGCACCCGUCGCUGUCGCGGCCGGUGGAUUUCCGAGGGCAGC ((((((...(((.(((.((((((((((..((((.....((((((((((....)))))...........((....)))))))....))))..)))...)))))))))).)))))))))... ( -43.50) >consensus GUCCUCGUAGGAUAUCUCAUCGGUGAUCUUGACCAAGAGCUCCAGCAGUUCCCUGCUAUUGCCGGCCGUCAUUGGAGGAGCUCCAGUCGC___CGCAGCCGGCGGAUUUCCGAGGACACC ((((((...(((.(((.(.(((((......(((...((((((((((((....)))))........(((....))).)))))))..)))((....)).))))).)))).)))))))))... (-37.70 = -38.45 + 0.75)

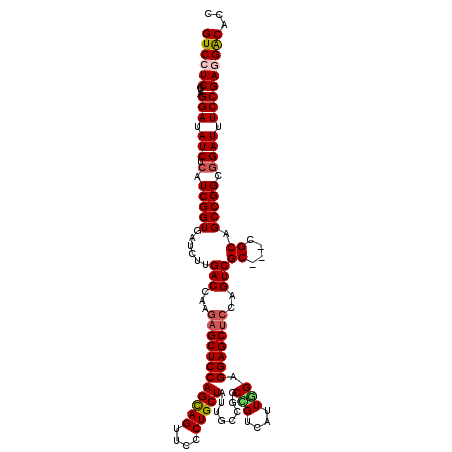

| Location | 8,403,568 – 8,403,685 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -45.20 |
| Consensus MFE | -31.66 |
| Energy contribution | -33.73 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8403568 117 - 22224390 GGUGUCAUCGGAAAUCCGCCGGCUGCG---GCGACUGGAGCUCCCCCAAUGACGGCCGCGAAUAGCAGGGAACUGCUGGAGCUCUUGGUCAAGAUAACCGAUGAGAUAUCCUAUGAGGAC .((.(((((((..(((.((((....))---))((((((((((((.((......))........(((((....)))))))))))).)))))..)))..))))))).)).(((.....))). ( -50.00) >DroSec_CAF1 2774 117 - 1 GGUGUCAUCGGAAAUCCGCCGGCUGCG---GCGACUGGAGCUCCUCCAAUGACAGCCGCUAAUAGCAGAGAACUGCUGGAGCUCUUGGUCAAGAUCACCGAUGAGAUAUCCUACGAGGAC .((.(((((((..(((.((((....))---))((((((((((((((....))...........(((((....)))))))))))).)))))..)))..))))))).)).(((.....))). ( -45.60) >DroEre_CAF1 6221 117 - 1 GCUGCCCUCGGAAAUCAGCCGGCCGCG---GCGACGGGAGCUCCCCAAAUGACCGCCGGCAACAGCAGGGAACUGCUGGAGCUCUUGGUCAAGAUCACCGAUGAGAUAUCCUACGAGGAC .....(((((((.(((.((((....))---))((((((.....)))...((((((..(((..((((((....))))))..)))..)))))).).))........))).)))...)))).. ( -47.60) >DroYak_CAF1 5859 120 - 1 GCUGCCCUCGGAAAUCCACCGGCCGCGACAGCGACGGGUGCUCCUCAAAUGACGGCCGGCAAUAGCAGGGAACUACUGGAGCUAUUGGUCAAAAUCACCGAUGAGAUAUCCUACGAGGAC .....(((((((.(((((((((((((....))((.((.....)))).......))))))(((((((.((......))...)))))))..............)).))).)))...)))).. ( -37.60) >consensus GCUGCCAUCGGAAAUCCGCCGGCCGCG___GCGACGGGAGCUCCCCAAAUGACGGCCGCCAAUAGCAGGGAACUGCUGGAGCUCUUGGUCAAGAUCACCGAUGAGAUAUCCUACGAGGAC ...(((((((((.(((((.(((.(((....))).((((((((((.....((.....)).....(((((....)))))))))))))))..........))).)).))).)))...)))))) (-31.66 = -33.73 + 2.06)


| Location | 8,403,608 – 8,403,725 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.25 |
| Mean single sequence MFE | -50.28 |
| Consensus MFE | -39.46 |
| Energy contribution | -39.90 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8403608 117 - 22224390 CACUGGGACACCAGUUGCUUCGACGGGAGCCCCGUCAACAGGUGUCAUCGGAAAUCCGCCGGCUGCG---GCGACUGGAGCUCCCCCAAUGACGGCCGCGAAUAGCAGGGAACUGCUGGA ..((((....))))((((...((((((...))))))....((((((((.((.....(((((....))---)))...((....)).)).))))).))))))).((((((....)))))).. ( -49.80) >DroSec_CAF1 2814 117 - 1 CACUGGGACACCAGUUGCUUCAACGGGAGCUCCGUCAAUAGGUGUCAUCGGAAAUCCGCCGGCUGCG---GCGACUGGAGCUCCUCCAAUGACAGCCGCUAAUAGCAGAGAACUGCUGGA .(((((....))))).((((((..((((((((((((....((..((....))...))((((....))---))))).)))))))))....))).)))......((((((....)))))).. ( -47.50) >DroEre_CAF1 6261 117 - 1 CACUGGGGCACCUGUUGCUCUAACGGGCGCCCCGUCCUCCGCUGCCCUCGGAAAUCAGCCGGCCGCG---GCGACGGGAGCUCCCCAAAUGACCGCCGGCAACAGCAGGGAACUGCUGGA ....(((((.(((((((...))))))).)))))....((((.......)))).....((((((...(---....)(((.....)))........))))))..((((((....)))))).. ( -54.10) >DroYak_CAF1 5899 120 - 1 CACUGGAGCACCUGUUGCUCCAACGGGCGCCCCGUCCUCCGCUGCCCUCGGAAAUCCACCGGCCGCGACAGCGACGGGUGCUCCUCAAAUGACGGCCGGCAAUAGCAGGGAACUACUGGA ....(((((((((((((((.....(((((...)))))..(((.(((...((....))...))).)))..)))))))))))))))...........((.((....)).))........... ( -49.70) >consensus CACUGGGACACCAGUUGCUCCAACGGGAGCCCCGUCAACAGCUGCCAUCGGAAAUCCGCCGGCCGCG___GCGACGGGAGCUCCCCAAAUGACGGCCGCCAAUAGCAGGGAACUGCUGGA ....(((((.((((((((....(((((...))))).....((.(((..(((....)))..))).))....)))))))).)))))..................((((((....)))))).. (-39.46 = -39.90 + 0.44)


| Location | 8,403,685 – 8,403,805 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.75 |
| Mean single sequence MFE | -55.97 |
| Consensus MFE | -47.42 |
| Energy contribution | -47.92 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8403685 120 + 22224390 UGUUGACGGGGCUCCCGUCGAAGCAACUGGUGUCCCAGUGGGCAGGACCAACGCACCGGCGGGCAGGUUGCCCACAUAGGCGUCCAAUUGGUUGAGCGCCGUCAGCUUGCGCUCCACAUU .((((((((.((((((((((..((.(((((....)))))((......))...))..))))))...((.((((......)))).))........)))).)))))))).............. ( -51.30) >DroSec_CAF1 2891 120 + 1 UAUUGACGGAGCUCCCGUUGAAGCAACUGGUGUCCCAGUGGGCAGGACCAACGCGCCGGAGGGCAGGUUGCCCACAUAGGCGUCCAGUUGGUUGAGCGCCGUCAGCUUGCGCUCCACAUU ...((((((.((((........((.(((((....)))))..))..((((((((((((.(.((((.....)))).)...)))))...))))))))))).))))))((....))........ ( -52.70) >DroEre_CAF1 6338 120 + 1 GGAGGACGGGGCGCCCGUUAGAGCAACAGGUGCCCCAGUGGGCAGGACCAACGAUCCGGCGGGCAGGUUGCCCACAUAGGCGUCCAGUUGGCUGAGCGCCGUUAGCUUGCGCUCUUCAUU ((((..(((((((((.(((.....))).)))))))).(..(((.(((.......)))(((((((.....)))).....((((..(((....)))..))))))).)))..))..))))... ( -58.70) >DroYak_CAF1 5979 120 + 1 GGAGGACGGGGCGCCCGUUGGAGCAACAGGUGCUCCAGUGGGCAGGACCAGCGCUCCGGCGGGGAGGUUGCCCACAUAGGCGUCCAGUUGGCUGAGCGCCGUCAGCUUACGCUCCGCAUU ((((....(((((((...(((((((.....)))))))((((((((..((..(((....))).))...))))))))...))))))).((.((((((......)))))).)).))))..... ( -61.20) >consensus GGAGGACGGGGCGCCCGUUGAAGCAACAGGUGCCCCAGUGGGCAGGACCAACGCUCCGGCGGGCAGGUUGCCCACAUAGGCGUCCAGUUGGCUGAGCGCCGUCAGCUUGCGCUCCACAUU ....(((((((((((.(((.....))).)))))))).(((((((..(((..(((....)))....))))))))))...((((..(((....)))..)))))))(((....)))....... (-47.42 = -47.92 + 0.50)


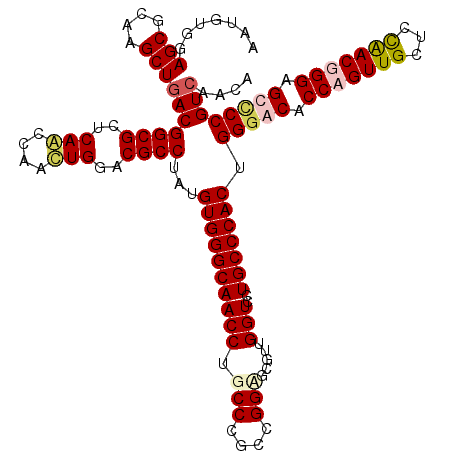
| Location | 8,403,685 – 8,403,805 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.75 |
| Mean single sequence MFE | -54.58 |
| Consensus MFE | -45.64 |
| Energy contribution | -47.07 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.973080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8403685 120 - 22224390 AAUGUGGAGCGCAAGCUGACGGCGCUCAACCAAUUGGACGCCUAUGUGGGCAACCUGCCCGCCGGUGCGUUGGUCCUGCCCACUGGGACACCAGUUGCUUCGACGGGAGCCCCGUCAACA .......(((....)))(((((.((((..((.....(((((((..((((((.....)))))).)).))))).(((..((..(((((....))))).))...))))))))).))))).... ( -53.20) >DroSec_CAF1 2891 120 - 1 AAUGUGGAGCGCAAGCUGACGGCGCUCAACCAACUGGACGCCUAUGUGGGCAACCUGCCCUCCGGCGCGUUGGUCCUGCCCACUGGGACACCAGUUGCUUCAACGGGAGCUCCGUCAAUA .......(((....)))(((((.((((...(((((((...((((.((((((((((((((....))))....)))..)))))))))))...)))))))((.....)))))).))))).... ( -55.40) >DroEre_CAF1 6338 120 - 1 AAUGAAGAGCGCAAGCUAACGGCGCUCAGCCAACUGGACGCCUAUGUGGGCAACCUGCCCGCCGGAUCGUUGGUCCUGCCCACUGGGGCACCUGUUGCUCUAACGGGCGCCCCGUCCUCC .......(((....)))...((((..(((....)))..))))...((((((.....)))))).((((.((.((.....)).)).(((((.(((((((...))))))).)))))))))... ( -54.90) >DroYak_CAF1 5979 120 - 1 AAUGCGGAGCGUAAGCUGACGGCGCUCAGCCAACUGGACGCCUAUGUGGGCAACCUCCCCGCCGGAGCGCUGGUCCUGCCCACUGGAGCACCUGUUGCUCCAACGGGCGCCCCGUCCUCC ...(((((((....)))...((((..(((....)))..)))).....((....))...)))).((((.((.((...(((((..(((((((.....)))))))..))))).)).)).)))) ( -54.80) >consensus AAUGUGGAGCGCAAGCUGACGGCGCUCAACCAACUGGACGCCUAUGUGGGCAACCUGCCCGCCGGAGCGUUGGUCCUGCCCACUGGGACACCAGUUGCUCCAACGGGAGCCCCGUCAACA .......(((....)))(((((((..(((....)))..))))...((((((((((.(((....))).....)))..))))))).(((((.(((((((...))))))).)))))))).... (-45.64 = -47.07 + 1.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:46 2006