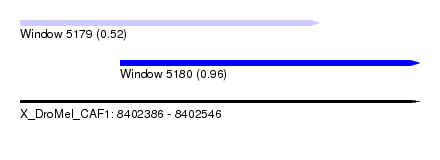
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,402,386 – 8,402,546 |
| Length | 160 |
| Max. P | 0.963331 |

| Location | 8,402,386 – 8,402,506 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -31.76 |
| Consensus MFE | -30.38 |
| Energy contribution | -30.42 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.96 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8402386 120 + 22224390 GUAAUCCUCAAGCGAUCCAAGCAUUAUCUCCAGCUGAUCCUCGCGCAGCACUAUGUGCUUGGCAAUGGCCGUUAAACUAUAGAUGGCCUUCAGCCGCACAUCCUCGAUCUCAUCGUUGAA ...........((((....(((..........))).....)))).((((...((((((..(((...(((((((........)))))))....)))))))))....(((...))))))).. ( -32.20) >DroSec_CAF1 1592 120 + 1 GUAAUCCUCAAGCGAUCCAAGCAUUAUCUCCAGCUGAUCCUCGCGCAGCACUAUGUGCCUGGCAAUGGCCGUUAAACUAUAGAUGGCCUUCAGCCGCACAUCCUCGAUCUCGUCGUUGAA ...........((((....(((..........))).....)))).((((...((((((..(((...(((((((........)))))))....)))))))))...((....))..)))).. ( -32.50) >DroSim_CAF1 5393 120 + 1 GUAAUCCUCAAGCGAUCCAAGCAUUAUCUCCAGCUGAUCCUCGCGCAGCACUAUGUGCCUGGCAAUGGCCGUUAAACUAUAGAUGGCCUUCAGCCGCACAUCCUCGAUCUCGUCGUUGAA ...........((((....(((..........))).....)))).((((...((((((..(((...(((((((........)))))))....)))))))))...((....))..)))).. ( -32.50) >DroEre_CAF1 5039 120 + 1 GUAAUCCUCAAGCGAACCAAGCAUUAUCUCCAGCUGAUCCUCGCGCAGCACUAUGUGCCUGGCAAUGGCCGUCAAACUGUAGAUGGCCUUCAAUCGCACAUCCUCGAUCUCGUCGUUAAA ...........((((.(((.((((........((((.........)))).....)))).)))....(((((((........))))))).....)))).......((((...))))..... ( -29.42) >DroYak_CAF1 4677 120 + 1 GUAAUCCUCCAGCGAACCAAGCAUUAUCUCCAGCUGAUCCUCGCGCAGCACUAUGUGCCUGGCAAUGGCCGUCAAGCUGUAGAUGGCCUUCAACCGCACAUCCUCGAUCUCGUCGUUGAA .........((((((.....((...((((.((((((((....(((((......)))))..(((....)))))).))))).))))((.......)))).......((....)))))))).. ( -32.20) >consensus GUAAUCCUCAAGCGAUCCAAGCAUUAUCUCCAGCUGAUCCUCGCGCAGCACUAUGUGCCUGGCAAUGGCCGUUAAACUAUAGAUGGCCUUCAGCCGCACAUCCUCGAUCUCGUCGUUGAA ...........((((....(((..........))).....)))).((((...((((((..(((...(((((((........)))))))....)))))))))....(((...))))))).. (-30.38 = -30.42 + 0.04)


| Location | 8,402,426 – 8,402,546 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -38.32 |
| Consensus MFE | -34.88 |
| Energy contribution | -35.12 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8402426 120 + 22224390 UCGCGCAGCACUAUGUGCUUGGCAAUGGCCGUUAAACUAUAGAUGGCCUUCAGCCGCACAUCCUCGAUCUCAUCGUUGAACAUGUCCACAAGGAAAUCUAGGCUGGUCACCGCGAAAUCC (((((.(((((...)))))((((...(((((((........)))))))..(((((...(((..(((((......)))))..)))(((....)))......))))))))).)))))..... ( -39.20) >DroSec_CAF1 1632 120 + 1 UCGCGCAGCACUAUGUGCCUGGCAAUGGCCGUUAAACUAUAGAUGGCCUUCAGCCGCACAUCCUCGAUCUCGUCGUUGAACAUAUCCACAAGGAAGUCCAGGCUGGUCACCGCGAAAUCC (((((((((...((((((..(((...(((((((........)))))))....))))))))).((.((..((.(.((.((.....)).)).).))..)).)))))).....)))))..... ( -38.50) >DroSim_CAF1 5433 120 + 1 UCGCGCAGCACUAUGUGCCUGGCAAUGGCCGUUAAACUAUAGAUGGCCUUCAGCCGCACAUCCUCGAUCUCGUCGUUGAACAUAUCCACAAGGAAGUCCAGGCUGGUCACCGCGAAAUCC (((((((((...((((((..(((...(((((((........)))))))....))))))))).((.((..((.(.((.((.....)).)).).))..)).)))))).....)))))..... ( -38.50) >DroEre_CAF1 5079 120 + 1 UCGCGCAGCACUAUGUGCCUGGCAAUGGCCGUCAAACUGUAGAUGGCCUUCAAUCGCACAUCCUCGAUCUCGUCGUUAAACAUGUCGACAAGGAAGUCCAGGCUGGUCACCGCAAAAUCC ..(((..(.((((...(((((((...(((((((........)))))))............((((.(...(((.(((.....))).)))).)))).).)))))))))))..)))....... ( -36.10) >DroYak_CAF1 4717 120 + 1 UCGCGCAGCACUAUGUGCCUGGCAAUGGCCGUCAAGCUGUAGAUGGCCUUCAACCGCACAUCCUCGAUCUCGUCGUUGAACAUGUCCACAAGGAAGUCCAGGCUGGUCACCGCGAAAUCC (((((..(.((((...(((((((...(((((((........)))))))((((((.((..(((...)))...)).))))))....(((....))).).)))))))))))..)))))..... ( -39.30) >consensus UCGCGCAGCACUAUGUGCCUGGCAAUGGCCGUUAAACUAUAGAUGGCCUUCAGCCGCACAUCCUCGAUCUCGUCGUUGAACAUGUCCACAAGGAAGUCCAGGCUGGUCACCGCGAAAUCC (((((((((...((((((..(((...(((((((........)))))))....))))))))).((.(((.((.(.((.((.....)).)).).)).))).)))))).....)))))..... (-34.88 = -35.12 + 0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:34 2006