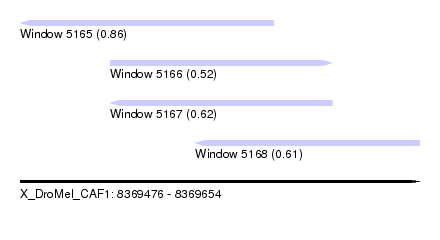
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,369,476 – 8,369,654 |
| Length | 178 |
| Max. P | 0.862344 |

| Location | 8,369,476 – 8,369,589 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.85 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -18.74 |
| Energy contribution | -18.74 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8369476 113 - 22224390 UAG-----CAGUGUAAUUUCUCGCCCUCAUGCCAAUGCAUUAAAUGCCAAUAAAUUCUAUAAAUUAAGAAUGGGCAU--UGCUACCAGCAUUUCGACAUAACUCAAUCGAGAAAACUGUU .((-----((((....(((((((.....((((....(((....(((((.....(((((........))))).)))))--))).....))))...((......))...))))))))))))) ( -24.80) >DroSec_CAF1 34609 113 - 1 UAG-----CAGUGUAAUUUCUCGCCCUCAUGCCAACGCAUUAAAUGCCAAUAAAUUCUAUAAAUUAAGAAUGGGCAU--UGCUACCAGCAUUUCCACAUAACUCAAUCGGGAAAACUGUU .((-----((((....((((((((((..((((....)))).............(((((........)))))))))..--(((.....)))..................)))))))))))) ( -24.00) >DroSim_CAF1 37828 113 - 1 UAG-----CAGUGUAAUUUCUCGCCCUCAUGCCAACGCAUUAAAUGCCAAUAAAUUCUAUAAAUUAAGAAUGGGCAU--UGCUACCAGCAUUUCCACAUAACUCAAUCGGGAAAACUGUU .((-----((((....((((((((((..((((....)))).............(((((........)))))))))..--(((.....)))..................)))))))))))) ( -24.00) >DroEre_CAF1 31269 117 - 1 UAGUAGUGUAAUGUAAUUUCUCGCCCUCAUGCCAACGCAUUAAAUGCCAAUAAAUUCUAUAAAUUAAGAAUGGGCA---UGCUACCAGCAUUUUCACAUUACUUAAUCGAGGAAACUGUU .((((((((.............((((..((((....)))).............(((((........)))))))))(---(((.....))))....))))))))......((....))... ( -25.00) >DroYak_CAF1 33694 112 - 1 --------UAAUGUAAUUUCUCGCCCUCAUGCCAACGCAUUAAAUGCCAAUAAAUUCGGUGAAUUAAGAAUGGGCAUUGUGCUACCAGCAUUUUCACAUUACUCAAUCGUGGAAACUGUU --------............................((((..((((((.....((((..........)))).))))))))))....((((((((((((((....))).))))))).)))) ( -19.20) >consensus UAG_____CAGUGUAAUUUCUCGCCCUCAUGCCAACGCAUUAAAUGCCAAUAAAUUCUAUAAAUUAAGAAUGGGCAU__UGCUACCAGCAUUUCCACAUAACUCAAUCGAGAAAACUGUU ................((((((((((..((((....)))).............(((((........)))))))))....(((.....)))..................))))))...... (-18.74 = -18.74 + 0.00)



| Location | 8,369,516 – 8,369,615 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.64 |
| Mean single sequence MFE | -26.29 |
| Consensus MFE | -13.12 |
| Energy contribution | -13.32 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.50 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8369516 99 + 22224390 A--AUGCCCAUUCUUAAUUUAUAGAAUUUAUUGGCAUUUAAUGCAUUGGCAUGAGGGCGAGAAAUUACACUG-----CUAUG--------------CAGCAUAAAUGCGCCAAUAAAAAA .--.......((((........))))(((((((((......(((((.((((.(...(..(....)..).)))-----)))))--------------))(((....))))))))))))... ( -24.20) >DroSec_CAF1 34649 111 + 1 A--AUGCCCAUUCUUAAUUUAUAGAAUUUAUUGGCAUUUAAUGCGUUGGCAUGAGGGCGAGAAAUUACACUG-----CUAUGCUCCGUCGCAUUUGCAGCAUAAAUGCGCCAAUAAAA-- .--.......((((........))))(((((((((....((((((..((((((.(((..(....)..).)).-----.))))))....)))))).(((.......)))))))))))).-- ( -29.60) >DroSim_CAF1 37868 111 + 1 A--AUGCCCAUUCUUAAUUUAUAGAAUUUAUUGGCAUUUAAUGCGUUGGCAUGAGGGCGAGAAAUUACACUG-----CUAUGCUCCGUCGCAUUUACAGCAUAAAUGCGCCAAUAAAA-- .--.......((((........))))(((((((((....((((((..((((((.(((..(....)..).)).-----.))))))....))))))....(((....)))))))))))).-- ( -29.30) >DroEre_CAF1 31309 108 + 1 A---UGCCCAUUCUUAAUUUAUAGAAUUUAUUGGCAUUUAAUGCGUUGGCAUGAGGGCGAGAAAUUACAUUACACUACUAUGCU-----GCCAUUGCAGUAUAAAUG--CCGAUAAAA-- .---......((((........))))((((((((((((((.((((.(((((.....(((((................)).))))-----)))).))))...))))))--)))))))).-- ( -28.59) >DroYak_CAF1 33734 101 + 1 ACAAUGCCCAUUCUUAAUUCACCGAAUUUAUUGGCAUUUAAUGCGUUGGCAUGAGGGCGAGAAAUUACAUUA----------CU-----ACCAUUGCGGCAUAAAUG--GCAAUAAAA-- ..........................(((((((.((((((.((((.(((.......(..(....)..)....----------..-----.))).))))...))))))--.))))))).-- ( -19.74) >consensus A__AUGCCCAUUCUUAAUUUAUAGAAUUUAUUGGCAUUUAAUGCGUUGGCAUGAGGGCGAGAAAUUACACUG_____CUAUGCU_____GCAAUUGCAGCAUAAAUGCGCCAAUAAAA__ ....((((((((((........))))).............((((....))))..)))))............................................................. (-13.12 = -13.32 + 0.20)

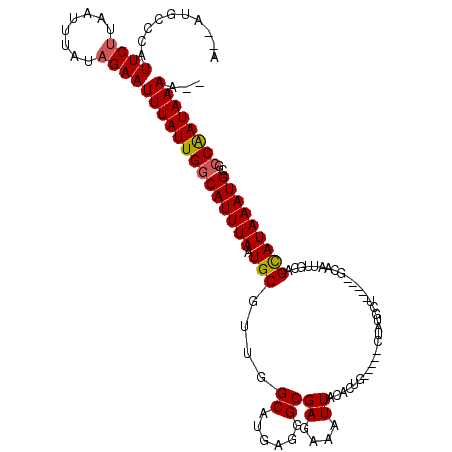
| Location | 8,369,516 – 8,369,615 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.64 |
| Mean single sequence MFE | -26.74 |
| Consensus MFE | -12.58 |
| Energy contribution | -12.98 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8369516 99 - 22224390 UUUUUUAUUGGCGCAUUUAUGCUG--------------CAUAG-----CAGUGUAAUUUCUCGCCCUCAUGCCAAUGCAUUAAAUGCCAAUAAAUUCUAUAAAUUAAGAAUGGGCAU--U .........((((...((((((((--------------(...)-----)))))))).....))))...((((....))))..((((((.....(((((........))))).)))))--) ( -25.50) >DroSec_CAF1 34649 111 - 1 --UUUUAUUGGCGCAUUUAUGCUGCAAAUGCGACGGAGCAUAG-----CAGUGUAAUUUCUCGCCCUCAUGCCAACGCAUUAAAUGCCAAUAAAUUCUAUAAAUUAAGAAUGGGCAU--U --.......((((...(((((((((..((((......)))).)-----)))))))).....))))...((((....))))..((((((.....(((((........))))).)))))--) ( -31.50) >DroSim_CAF1 37868 111 - 1 --UUUUAUUGGCGCAUUUAUGCUGUAAAUGCGACGGAGCAUAG-----CAGUGUAAUUUCUCGCCCUCAUGCCAACGCAUUAAAUGCCAAUAAAUUCUAUAAAUUAAGAAUGGGCAU--U --.......((((...(((((((((..((((......)))).)-----)))))))).....))))...((((....))))..((((((.....(((((........))))).)))))--) ( -29.90) >DroEre_CAF1 31309 108 - 1 --UUUUAUCGG--CAUUUAUACUGCAAUGGC-----AGCAUAGUAGUGUAAUGUAAUUUCUCGCCCUCAUGCCAACGCAUUAAAUGCCAAUAAAUUCUAUAAAUUAAGAAUGGGCA---U --........(--((((.(((((((.(((..-----..))).))))))))))))........((((..((((....)))).............(((((........))))))))).---. ( -25.10) >DroYak_CAF1 33734 101 - 1 --UUUUAUUGC--CAUUUAUGCCGCAAUGGU-----AG----------UAAUGUAAUUUCUCGCCCUCAUGCCAACGCAUUAAAUGCCAAUAAAUUCGGUGAAUUAAGAAUGGGCAUUGU --..(((((((--((((........))))))-----))----------)))...........((((...........(((..(((........)))..)))..........))))..... ( -21.70) >consensus __UUUUAUUGGCGCAUUUAUGCUGCAAAGGC_____AGCAUAG_____CAGUGUAAUUUCUCGCCCUCAUGCCAACGCAUUAAAUGCCAAUAAAUUCUAUAAAUUAAGAAUGGGCAU__U ................((((((((........................))))))))......((((..((((....)))).............(((((........)))))))))..... (-12.58 = -12.98 + 0.40)

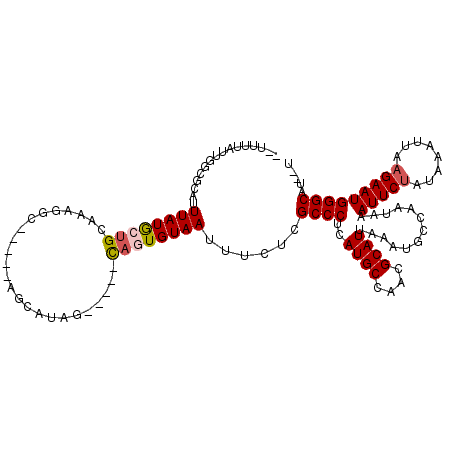

| Location | 8,369,554 – 8,369,654 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.09 |
| Mean single sequence MFE | -24.06 |
| Consensus MFE | -6.04 |
| Energy contribution | -7.24 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.25 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8369554 100 - 22224390 CUAAUUUCC-CCGCAGUGUGGAAAAUUCAUUUUUCUUUUUUUUUUUAUUGGCGCAUUUAUGCUG--------------CAUAG-----CAGUGUAAUUUCUCGCCCUCAUGCCAAUGCAU .........-..(((((((((((((.....)))))..............((((...((((((((--------------(...)-----)))))))).....))))..)))))...))).. ( -22.40) >DroSec_CAF1 34687 104 - 1 CUAAUUCCC-CCGCGCCGUGGAAAACUCAUUU----------UUUUAUUGGCGCAUUUAUGCUGCAAAUGCGACGGAGCAUAG-----CAGUGUAAUUUCUCGCCCUCAUGCCAACGCAU .........-..(((((((((((((.....))----------))))).))))))..(((((((((..((((......)))).)-----))))))))............((((....)))) ( -29.20) >DroSim_CAF1 37906 104 - 1 CUAAUUUCC-CCGCGCCGUGGAAAACUCAUUU----------UUUUAUUGGCGCAUUUAUGCUGUAAAUGCGACGGAGCAUAG-----CAGUGUAAUUUCUCGCCCUCAUGCCAACGCAU .........-..(((((((((((((.....))----------))))).))))))..(((((((((..((((......)))).)-----))))))))............((((....)))) ( -27.60) >DroEre_CAF1 31346 101 - 1 CUAAUUUGC-CCGAGCUGUGGAAAAUUCA-UU----------UUUUAUCGG--CAUUUAUACUGCAAUGGC-----AGCAUAGUAGUGUAAUGUAAUUUCUCGCCCUCAUGCCAACGCAU .........-.((((((((((((((....-))----------))))).)))--)..(((((((((.(((..-----..))).))))))))).........))).....((((....)))) ( -21.00) >DroYak_CAF1 33774 93 - 1 CUAAUUUCCACCGAGCUGUGGAAAAUUCAAUU----------UUUUAUUGC--CAUUUAUGCCGCAAUGGU-----AG----------UAAUGUAAUUUCUCGCCCUCAUGCCAACGCAU ............(((..(((..(((((((...----------..(((((((--((((........))))))-----))----------))))).)))))..))).)))((((....)))) ( -20.10) >consensus CUAAUUUCC_CCGCGCUGUGGAAAAUUCAUUU__________UUUUAUUGGCGCAUUUAUGCUGCAAAGGC_____AGCAUAG_____CAGUGUAAUUUCUCGCCCUCAUGCCAACGCAU ..........(((.....)))............................((((...((((((((........................)))))))).....))))...((((....)))) ( -6.04 = -7.24 + 1.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:23 2006