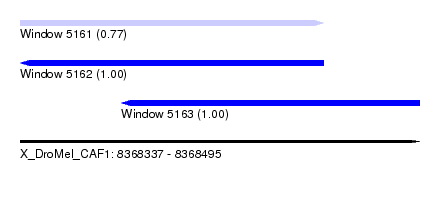
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,368,337 – 8,368,495 |
| Length | 158 |
| Max. P | 0.999282 |

| Location | 8,368,337 – 8,368,457 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.54 |
| Mean single sequence MFE | -26.33 |
| Consensus MFE | -18.48 |
| Energy contribution | -18.80 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8368337 120 + 22224390 UUAGUUUGAUGAAUAUCUUAUACUUCUUUUUUUUUUUUUUUAGUUUUUUGUUCGUGUUGUGGGGCUACUAAACUUCCGUUUAGUGCGUUAGUUGGCCCGCUUUAAAUGUGGUUAAACUGA (((((((((((((((......(((.................)))....))))).((..(((((.(.(((((((....)))))))((....)).).)))))..))......)))))))))) ( -24.83) >DroSec_CAF1 33491 109 + 1 UUAGUUUGAUGAAUAUCUUAUUCUUAUU----UUAU-------UUUUUCGGUCGUGUUGUGGGGCCACUAAACUUCCGUUUAGUGCGUUAGUUGGCCCGCUUUAAAUGUGGUUAAGCUGA (((((((((((((((...))))).....----....-------.........((..((..(((((((....(((..(((.....)))..))))))))).)....))..)))))))))))) ( -26.90) >DroSim_CAF1 36696 113 + 1 UUAGUUUGAUGAAUAUCUUAUUCUUAUUUGUUUUUU-------UUUUUCGGUCGUGUUGUGGGGCCACUAAACUUCCGUUUAGUGCGUUAGUUGGCCCGCUUUAAAUGUGGUUAAACUGA (((((((((((((((...))))).............-------.........((..((..(((((((....(((..(((.....)))..))))))))).)....))..)))))))))))) ( -26.90) >DroEre_CAF1 30162 96 + 1 UUACUGUGAUGAAUAUUCC--------U----CAUU-------UUUAGCC---GUGUUGCG-GGCC-UAAAAGUUUCGUUUAGUGCGCUAGUUGGCCCGCUUUAAAUGUGGUUAAGCUGA .....((((.((....)).--------)----))).-------(((((((---(..(((((-((((-....((...(((.....)))))....)))))))....))..)))))))).... ( -27.20) >DroYak_CAF1 32466 100 + 1 UUAGUUUGAUGAGUAUCCU--------U----UUGU-------UUUGGCGUUGGUGUUGUGGGGCC-CAAAACUUCCGCUUAGUGCGUUAGUUGGCCCGCUUUAAAUGUGGUUAAAUGGA ...............(((.--------(----(((.-------..(.(((((......(((((.((-...((((..(((.....)))..)))))))))))....))))).).)))).))) ( -25.80) >consensus UUAGUUUGAUGAAUAUCUUAU_CUU_UU____UUUU_______UUUUUCGGUCGUGUUGUGGGGCCACUAAACUUCCGUUUAGUGCGUUAGUUGGCCCGCUUUAAAUGUGGUUAAACUGA ((((((((((...................................................(((((....((((..(((.....)))..)))))))))((.......)).)))))))))) (-18.48 = -18.80 + 0.32)


| Location | 8,368,337 – 8,368,457 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.54 |
| Mean single sequence MFE | -20.93 |
| Consensus MFE | -11.86 |
| Energy contribution | -13.30 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.57 |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.998220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8368337 120 - 22224390 UCAGUUUAACCACAUUUAAAGCGGGCCAACUAACGCACUAAACGGAAGUUUAGUAGCCCCACAACACGAACAAAAAACUAAAAAAAAAAAAAAAGAAGUAUAAGAUAUUCAUCAAACUAA ..(((((.............(.((((...(....).(((((((....))))))).)))))...........................................(((....)))))))).. ( -18.80) >DroSec_CAF1 33491 109 - 1 UCAGCUUAACCACAUUUAAAGCGGGCCAACUAACGCACUAAACGGAAGUUUAGUGGCCCCACAACACGACCGAAAAA-------AUAA----AAUAAGAAUAAGAUAUUCAUCAAACUAA ((.((((...........))))(((((..(....).(((((((....))))))))))))............))....-------....----.....(((((...))))).......... ( -23.60) >DroSim_CAF1 36696 113 - 1 UCAGUUUAACCACAUUUAAAGCGGGCCAACUAACGCACUAAACGGAAGUUUAGUGGCCCCACAACACGACCGAAAAA-------AAAAAACAAAUAAGAAUAAGAUAUUCAUCAAACUAA ..(((((.............(.(((((..(....).(((((((....))))))))))))).................-------.............(((((...)))))...))))).. ( -24.10) >DroEre_CAF1 30162 96 - 1 UCAGCUUAACCACAUUUAAAGCGGGCCAACUAGCGCACUAAACGAAACUUUUA-GGCC-CGCAACAC---GGCUAAA-------AAUG----A--------GGAAUAUUCAUCACAGUAA ....................(((((((......((.......)).........-))))-))).....---.(((...-------.(((----(--------(.....)))))...))).. ( -17.86) >DroYak_CAF1 32466 100 - 1 UCCAUUUAACCACAUUUAAAGCGGGCCAACUAACGCACUAAGCGGAAGUUUUG-GGCCCCACAACACCAACGCCAAA-------ACAA----A--------AGGAUACUCAUCAAACUAA (((.(((((......)))))(.(((((((((..(((.....)))..))))...-)))))).................-------....----.--------.)))............... ( -20.30) >consensus UCAGUUUAACCACAUUUAAAGCGGGCCAACUAACGCACUAAACGGAAGUUUAGUGGCCCCACAACACGAACGAAAAA_______AAAA____AA_AAG_AUAAGAUAUUCAUCAAACUAA ......................(((((..(....).(((((((....))))))))))))............................................................. (-11.86 = -13.30 + 1.44)


| Location | 8,368,377 – 8,368,495 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.26 |
| Mean single sequence MFE | -23.85 |
| Consensus MFE | -11.86 |
| Energy contribution | -13.30 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.50 |
| SVM decision value | 3.48 |
| SVM RNA-class probability | 0.999282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8368377 118 - 22224390 UAAUAUCCACACUAUAUCCACAACAUACGCUAG--UAUCCUCAGUUUAACCACAUUUAAAGCGGGCCAACUAACGCACUAAACGGAAGUUUAGUAGCCCCACAACACGAACAAAAAACUA ........................((((....)--)))....(((((...............((((...(....).(((((((....))))))).))))...............))))). ( -18.11) >DroSec_CAF1 33523 106 - 1 ---------CACUAUAUCCCCACCAUACGCUAG--UAUCCUCAGCUUAACCACAUUUAAAGCGGGCCAACUAACGCACUAAACGGAAGUUUAGUGGCCCCACAACACGACCGAAAAA--- ---------...............((((....)--)))..((.((((...........))))(((((..(....).(((((((....))))))))))))............))....--- ( -23.40) >DroSim_CAF1 36732 108 - 1 ---------CACCAUAUCCCCACCAUACGCUAGUGUAUCCUCAGUUUAACCACAUUUAAAGCGGGCCAACUAACGCACUAAACGGAAGUUUAGUGGCCCCACAACACGACCGAAAAA--- ---------..................((...((((.......((((...........))))(((((..(....).(((((((....))))))))))))....))))...)).....--- ( -25.20) >DroEre_CAF1 30186 98 - 1 ----------A-CAUGGCCCCACCAUAUA---GCAUAUCCUCAGCUUAACCACAUUUAAAGCGGGCCAACUAGCGCACUAAACGAAACUUUUA-GGCC-CGCAACAC---GGCUAAA--- ----------.-..(((((.........(---((.........)))..............(((((((......((.......)).........-))))-))).....---)))))..--- ( -22.56) >DroYak_CAF1 32490 106 - 1 UGAUAU----G-CAUAGCCAC---AUAUAU--GCAUAUCCUCCAUUUAACCACAUUUAAAGCGGGCCAACUAACGCACUAAGCGGAAGUUUUG-GGCCCCACAACACCAACGCCAAA--- .(((((----(-((((.....---...)))--))))))).....................(.(((((((((..(((.....)))..))))...-)))))).................--- ( -30.00) >consensus _________CACCAUAUCCCCACCAUACGCUAG__UAUCCUCAGUUUAACCACAUUUAAAGCGGGCCAACUAACGCACUAAACGGAAGUUUAGUGGCCCCACAACACGAACGAAAAA___ ..............................................................(((((..(....).(((((((....))))))))))))..................... (-11.86 = -13.30 + 1.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:18 2006