
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 995,122 – 995,213 |
| Length | 91 |
| Max. P | 0.959862 |

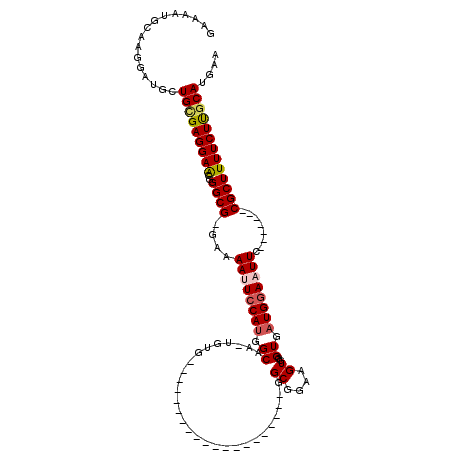
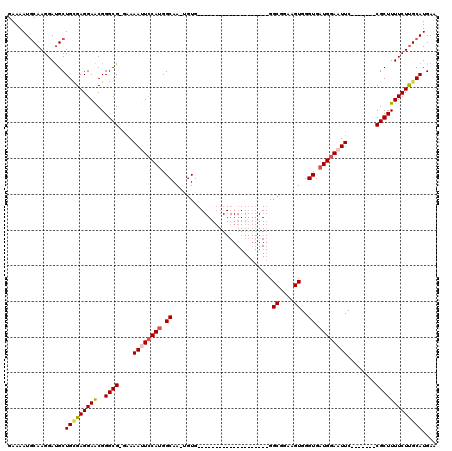
| Location | 995,122 – 995,213 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.38 |
| Mean single sequence MFE | -32.04 |
| Consensus MFE | -18.95 |
| Energy contribution | -19.51 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955116 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 995122 91 + 22224390 GAAAAUGCAAGGAUGCUGCGAGGAACGGGCG-GGAAAUUCCAUGGCAA-UGUG--------------------GGCGGAAGUGGGUGAUGGAAUUC-------CGCUUUUCUUGCAUGAA ......(((....)))(((((((((..((((-(((..((((((.((..-....--------------------.((....))..)).)))))))))-------))))))))))))).... ( -32.30) >DroSec_CAF1 77669 92 + 1 GAAAAUGCAAGGAUGCUGCGAGGAACGGGCG-GAAAAUUCCAUGGCAAAAGUG--------------------GGCGGAAGUGGGUGAUGGAAUUC-------CGCUUUUCUUGCAUGAA ......(((....)))(((((((((..((((-((..(((((((.((.......--------------------.((....))..)).)))))))))-------))))))))))))).... ( -32.40) >DroSim_CAF1 57169 92 + 1 GAAAAUGCAAGGAUGCUGCGAGGAACGGGCG-GAAAAUUCCAUGGCAAAAGUG--------------------GGCGGAAGUGGGUGAUGGAAUUC-------CGCUUUUCUUGCAUGAA ......(((....)))(((((((((..((((-((..(((((((.((.......--------------------.((....))..)).)))))))))-------))))))))))))).... ( -32.40) >DroEre_CAF1 91187 92 + 1 GAAAAUCCAAGGAUGCUGAGAGGAACGGGCGGAAAAACUGCAUGGCAA-UGUG--------------------GGCGGAAGUGGGUGAUGGAAUUC-------CGCUUUUCUCGCAUGAA ............((((.((((((....(((((((...(..(((....)-))..--------------------)((....))...........)))-------))))))))))))))... ( -27.60) >DroYak_CAF1 84138 118 + 1 GAAAAUGCAAGGACGCUGUGAGGAGCGGGCG-AAAAACUCCACGGCAA-UGUGGACGGAAGUGGGUGGAAGUGGGCGGAAGUGGGUGAUGGAAUUCCUUGUCUCGCUUUUCUUGCAUGAA ....(((((((((((((......))))((((-(...(((((((..(..-...).((....))..)))).)))..((....)).((..(.((....)))..))))))).)))))))))... ( -35.50) >consensus GAAAAUGCAAGGAUGCUGCGAGGAACGGGCG_GAAAAUUCCAUGGCAA_UGUG____________________GGCGGAAGUGGGUGAUGGAAUUC_______CGCUUUUCUUGCAUGAA ................(((((((((..((((....((((((((.((............................((....))..)).))))))))........))))))))))))).... (-18.95 = -19.51 + 0.56)

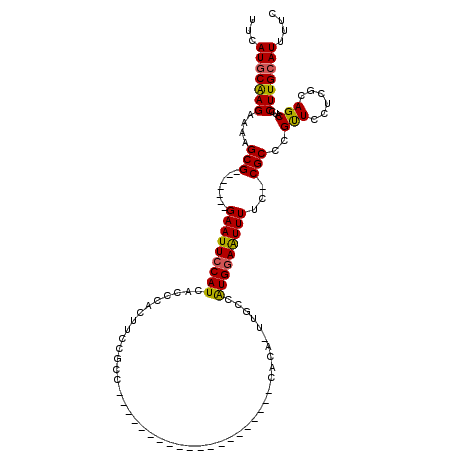
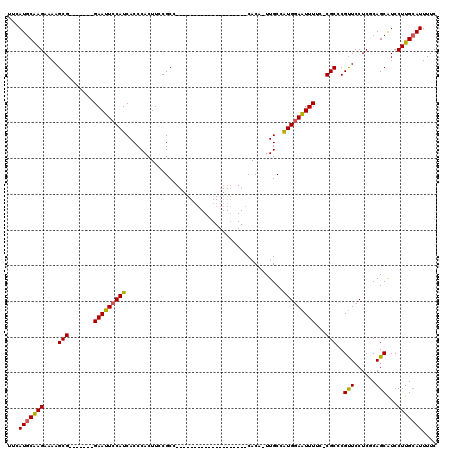
| Location | 995,122 – 995,213 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.38 |
| Mean single sequence MFE | -21.88 |
| Consensus MFE | -14.02 |
| Energy contribution | -13.70 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 995122 91 - 22224390 UUCAUGCAAGAAAAGCG-------GAAUUCCAUCACCCACUUCCGCC--------------------CACA-UUGCCAUGGAAUUUCC-CGCCCGUUCCUCGCAGCAUCCUUGCAUUUUC ...(((((((....(((-------(((.............)))))).--------------------....-((((...(((((....-.....)))))..))))....))))))).... ( -21.82) >DroSec_CAF1 77669 92 - 1 UUCAUGCAAGAAAAGCG-------GAAUUCCAUCACCCACUUCCGCC--------------------CACUUUUGCCAUGGAAUUUUC-CGCCCGUUCCUCGCAGCAUCCUUGCAUUUUC ...(((((((....(((-------(((.............)))))).--------------------.....((((...(((((....-.....)))))..))))....))))))).... ( -21.72) >DroSim_CAF1 57169 92 - 1 UUCAUGCAAGAAAAGCG-------GAAUUCCAUCACCCACUUCCGCC--------------------CACUUUUGCCAUGGAAUUUUC-CGCCCGUUCCUCGCAGCAUCCUUGCAUUUUC ...(((((((....(((-------(((.............)))))).--------------------.....((((...(((((....-.....)))))..))))....))))))).... ( -21.72) >DroEre_CAF1 91187 92 - 1 UUCAUGCGAGAAAAGCG-------GAAUUCCAUCACCCACUUCCGCC--------------------CACA-UUGCCAUGCAGUUUUUCCGCCCGUUCCUCUCAGCAUCCUUGGAUUUUC .....(((.((((((((-------(((.............)))))).--------------------...(-((((...)))))))))))))....(((.............)))..... ( -16.94) >DroYak_CAF1 84138 118 - 1 UUCAUGCAAGAAAAGCGAGACAAGGAAUUCCAUCACCCACUUCCGCCCACUUCCACCCACUUCCGUCCACA-UUGCCGUGGAGUUUUU-CGCCCGCUCCUCACAGCGUCCUUGCAUUUUC ...(((((((....((((((...((((.............)))).....................(((((.-.....)))))...)))-))).((((......))))..))))))).... ( -27.22) >consensus UUCAUGCAAGAAAAGCG_______GAAUUCCAUCACCCACUUCCGCC____________________CACA_UUGCCAUGGAAUUUUC_CGCCCGUUCCUCGCAGCAUCCUUGCAUUUUC ...(((((((....(((.......(((((((((............................................)))))))))...)))..(((......)))...))))))).... (-14.02 = -13.70 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:31 2006