
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,367,623 – 8,367,857 |
| Length | 234 |
| Max. P | 0.985026 |

| Location | 8,367,623 – 8,367,720 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.93 |
| Mean single sequence MFE | -18.53 |
| Consensus MFE | -10.64 |
| Energy contribution | -10.24 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8367623 97 + 22224390 -------ACAACGCAAUAAAGUGAAAUUAUCUUGAAAACUUUAUCAUUAUUUA----------------CUUGUAUAUUUGCGAUCUACAGGCGAUCGCGUUUCAUUUUCUGCAAAUGCA -------.....(((..((((((((((.....(((........))).......----------------...........((((((.......)))))))))))))))).)))....... ( -19.00) >DroSec_CAF1 32828 96 + 1 -------UCAACUCAAUAAAGUU-AUAUAUCUAGAUAACUUUAUCUUUAUUUA----------------UUUGUAUCUUUGCGAUCUACAGACGAUCGCGUUUCAUUUUCCGCAUAUGCA -------........((((((((-((........)))))))))).........----------------...........((((((.......))))))............((....)). ( -17.20) >DroSim_CAF1 36011 97 + 1 -------UCAACUCAAUAAAGUGGAUAUAUCUUGAUAACUUUAUCUUUAUUUA----------------UUUGUAUCUUUGCGAUCUACAGACGAUCGCAUUUCAUUUUCCGCAUAUGCA -------..........((((((((((((....(((((........)))))..----------------..))))))..(((((((.......)))))))...))))))..((....)). ( -19.00) >DroEre_CAF1 29561 98 + 1 UAAACACACAACUCAAUAAAGUGGGUUUAUCUUUAU--CUUU----UUAUAUA----------------UUUGUAUUUAUGCGAUCUUCAGGCGAUCGUGUUUCAUUUUCCGCAGAUGCA .((((((((((....((((((.((((........))--))))----))))...----------------.))))........((((.......))))))))))........((....)). ( -14.20) >DroYak_CAF1 31783 114 + 1 UAAACAAACAACUCAAUAAAAUGGGUGUAUCUUUAU--CUUU----AUAUUUAUCUUGAUAAAUUUGUUUUUGCAUUUAUGCGAUCUUCAGGCGAUCGCAUUUCAUUUUUCGCAGAUGCA .(((((((..(((((......))))).....(((((--(...----...........))))))))))))).(((((((((((((((.......))))))))............))))))) ( -23.24) >consensus _______ACAACUCAAUAAAGUGGAUAUAUCUUGAUAACUUUAUC_UUAUUUA________________UUUGUAUCUUUGCGAUCUACAGGCGAUCGCGUUUCAUUUUCCGCAGAUGCA ...............................................................................(((((((.......)))))))...........((....)). (-10.64 = -10.24 + -0.40)


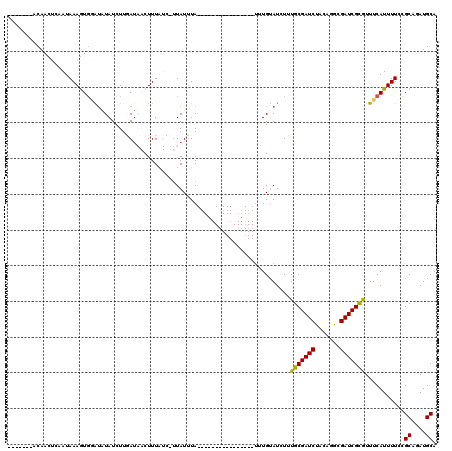
| Location | 8,367,623 – 8,367,720 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.93 |
| Mean single sequence MFE | -19.44 |
| Consensus MFE | -10.10 |
| Energy contribution | -10.14 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.609948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8367623 97 - 22224390 UGCAUUUGCAGAAAAUGAAACGCGAUCGCCUGUAGAUCGCAAAUAUACAAG----------------UAAAUAAUGAUAAAGUUUUCAAGAUAAUUUCACUUUAUUGCGUUGU------- .(((..((((((((.(((((.((((((.......))))))...........----------------.......(((........)))......))))).))).))))).)))------- ( -19.30) >DroSec_CAF1 32828 96 - 1 UGCAUAUGCGGAAAAUGAAACGCGAUCGUCUGUAGAUCGCAAAGAUACAAA----------------UAAAUAAAGAUAAAGUUAUCUAGAUAUAU-AACUUUAUUGAGUUGA------- .((....)).........(((((((((.......))))))...........----------------........(((((((((((........))-)))))))))..)))..------- ( -20.20) >DroSim_CAF1 36011 97 - 1 UGCAUAUGCGGAAAAUGAAAUGCGAUCGUCUGUAGAUCGCAAAGAUACAAA----------------UAAAUAAAGAUAAAGUUAUCAAGAUAUAUCCACUUUAUUGAGUUGA------- .((....))(((........(((((((.......)))))))..........----------------........((((....))))........)))((((....))))...------- ( -17.80) >DroEre_CAF1 29561 98 - 1 UGCAUCUGCGGAAAAUGAAACACGAUCGCCUGAAGAUCGCAUAAAUACAAA----------------UAUAUAA----AAAG--AUAAAGAUAAACCCACUUUAUUGAGUUGUGUGUUUA .((....)).............(((((.......))))).........(((----------------(((((((----...(--((((((.........)))))))...)))))))))). ( -18.30) >DroYak_CAF1 31783 114 - 1 UGCAUCUGCGAAAAAUGAAAUGCGAUCGCCUGAAGAUCGCAUAAAUGCAAAAACAAAUUUAUCAAGAUAAAUAU----AAAG--AUAAAGAUACACCCAUUUUAUUGAGUUGUUUGUUUA ((((((..........)).((((((((.......))))))))...)))).((((((((...((((...((((..----....--((....))......))))..))))...)))))))). ( -21.60) >consensus UGCAUAUGCGGAAAAUGAAACGCGAUCGCCUGUAGAUCGCAAAAAUACAAA________________UAAAUAA_GAUAAAGUUAUCAAGAUAAAUCCACUUUAUUGAGUUGU_______ (((....)))...........((((((.......))))))................................................................................ (-10.10 = -10.14 + 0.04)


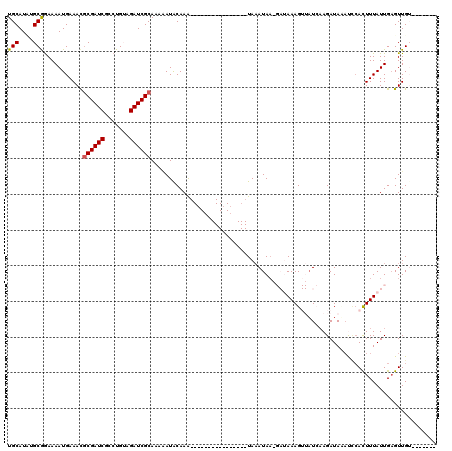
| Location | 8,367,656 – 8,367,757 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 83.39 |
| Mean single sequence MFE | -17.18 |
| Consensus MFE | -13.65 |
| Energy contribution | -14.03 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8367656 101 + 22224390 UUAUCAUUAUUUACUUGUAUAUUUGCGAUCUACAGGCGAUCGCGUUUCAUUUUCUGCAAAUGCAAACAAUUUUUUUUUGCCGGCAUUUACUCGUA--UUUGAU .....((((..(((..((......((((((.......))))))..............((((((...(((.......)))...))))))))..)))--..)))) ( -18.70) >DroSec_CAF1 32860 100 + 1 UUAUCUUUAUUUAUUUGUAUCUUUGCGAUCUACAGACGAUCGCGUUUCAUUUUCCGCAUAUGCAAACAGUUC---UUUGUCGGCAUUUACUCGUAUAUUUGAU ..........(((..(((((....((((((.......))))))................((((..((((...---.))))..))))......)))))..))). ( -17.50) >DroSim_CAF1 36044 100 + 1 UUAUCUUUAUUUAUUUGUAUCUUUGCGAUCUACAGACGAUCGCAUUUCAUUUUCCGCAUAUGCAAACAGUUC---UUUGUCGGCAUUUACUCGUAUAUUUAAU ..........(((..(((((...(((((((.......)))))))...............((((..((((...---.))))..))))......)))))..))). ( -18.10) >DroEre_CAF1 29599 88 + 1 UU----UUAUAUAUUUGUAUUUAUGCGAUCUUCAGGCGAUCGUGUUUCAUUUUCCGCAGAUGCAAACAGUUC---UUUGUCGGCAAUU--------AUUUGAU ..----(((.(((.((((......((((((.......))))))(((((((((.....))))).)))).....---.......)))).)--------)).))). ( -14.40) >consensus UUAUCUUUAUUUAUUUGUAUCUUUGCGAUCUACAGACGAUCGCGUUUCAUUUUCCGCAUAUGCAAACAGUUC___UUUGUCGGCAUUUACUCGUA_AUUUGAU .......................(((((((.......))))))).............((((((..((((.......))))..))))))............... (-13.65 = -14.03 + 0.38)


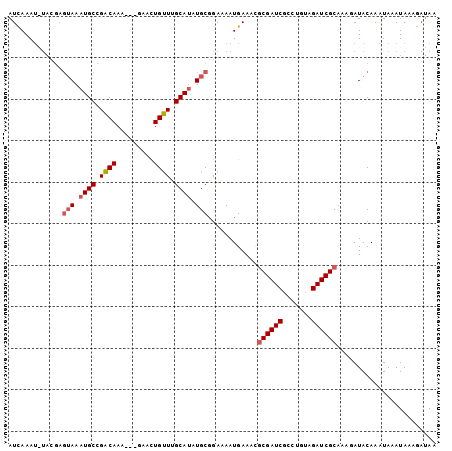
| Location | 8,367,656 – 8,367,757 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.39 |
| Mean single sequence MFE | -19.53 |
| Consensus MFE | -14.61 |
| Energy contribution | -15.43 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.985026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8367656 101 - 22224390 AUCAAA--UACGAGUAAAUGCCGGCAAAAAAAAAUUGUUUGCAUUUGCAGAAAAUGAAACGCGAUCGCCUGUAGAUCGCAAAUAUACAAGUAAAUAAUGAUAA ((((..--.....((((((((..((((.......))))..))))))))............((((((.......))))))..................)))).. ( -25.20) >DroSec_CAF1 32860 100 - 1 AUCAAAUAUACGAGUAAAUGCCGACAAA---GAACUGUUUGCAUAUGCGGAAAAUGAAACGCGAUCGUCUGUAGAUCGCAAAGAUACAAAUAAAUAAAGAUAA (((..........(((.((((.((((..---....)))).)))).)))............((((((.......))))))...))).................. ( -20.90) >DroSim_CAF1 36044 100 - 1 AUUAAAUAUACGAGUAAAUGCCGACAAA---GAACUGUUUGCAUAUGCGGAAAAUGAAAUGCGAUCGUCUGUAGAUCGCAAAGAUACAAAUAAAUAAAGAUAA .............(((.((((.((((..---....)))).)))).)))...........(((((((.......)))))))....................... ( -20.60) >DroEre_CAF1 29599 88 - 1 AUCAAAU--------AAUUGCCGACAAA---GAACUGUUUGCAUCUGCGGAAAAUGAAACACGAUCGCCUGAAGAUCGCAUAAAUACAAAUAUAUAA----AA .((....--------...(((.((((..---....)))).)))......))..........(((((.......)))))...................----.. ( -11.42) >consensus AUCAAAU_UACGAGUAAAUGCCGACAAA___GAACUGUUUGCAUAUGCGGAAAAUGAAACGCGAUCGCCUGUAGAUCGCAAAGAUACAAAUAAAUAAAGAUAA .............(((.((((.((((.........)))).)))).)))............((((((.......))))))........................ (-14.61 = -15.43 + 0.81)


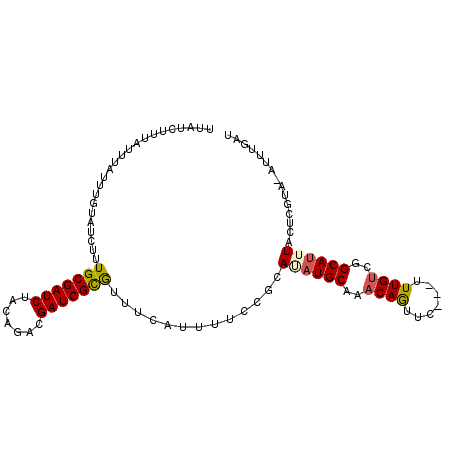
| Location | 8,367,757 – 8,367,857 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.20 |
| Mean single sequence MFE | -22.08 |
| Consensus MFE | -17.48 |
| Energy contribution | -17.76 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8367757 100 + 22224390 CGAAUUUAUCAUUUAGUUUGUUUCCAGUCUU---AAACAAACAAC------AAAUAAAGCUUCGUCGUU-UUAUGGAGCUUUU----------UUCCAGCCUCAAAUUGUGACUCAAAGC .((..((((.((((.((((((((........---))))))))...------....((((((((((....-..)))))))))).----------..........)))).)))).))..... ( -17.30) >DroSec_CAF1 32960 99 + 1 CGAAUUUGUCAUUUAGUUUGUUUCCAGCCUU---AAACAAACAAC------AAAUAAAGCUUCGUCGUU-UUAUGGAGCUUU-----------UUCCAGACUCAAAUGGUGACUCAAAGU .((..(..((((((.((((((((........---))))))))...------....((((((((((....-..))))))))))-----------..........))))))..).))..... ( -21.40) >DroSim_CAF1 36144 100 + 1 CGAAUUUGUCAUUUAGUUUGUUUCCAGCCUU---AAACAAACAAC------AAAUAAAGCUUCGUCGUU-UUGUGGAGCUUUU----------UUCCAGACUCAAACGGUGACUCAAAGU .......((((((..((((((((........---))))))))...------....(((((((((.....-...))))))))).----------..............))))))....... ( -19.70) >DroEre_CAF1 29687 100 + 1 CGAAUUUGUCAUUUAGUUUGUUGUCUGCCUU---AAACAAACAAC------AAAUAAAGCUUCGUCGUU-UUAUGGAGCUUUU----------UUCCAGACUCAAAUGGUGACUCAAAAC .((..(..((((((.(((((((((.((....---...)).)))))------))))((((((((((....-..)))))))))).----------..........))))))..).))..... ( -23.80) >DroYak_CAF1 31926 120 + 1 CGAGUUUGUCAUUUAGUUUGUUUUCUGCCUUUUCAAACAAACAGCCAAUAAAAAUAAAGCUUCGUCGUUUUUAUGAAGCUUUUCUGUUUCUGUUUCCAGACUCAAAUGGUGACUCAAAUC .(((((.........((((((((...........)))))))).((((........((((((((((.......))))))))))..((..((((....))))..))..)))))))))..... ( -28.20) >consensus CGAAUUUGUCAUUUAGUUUGUUUCCAGCCUU___AAACAAACAAC______AAAUAAAGCUUCGUCGUU_UUAUGGAGCUUUU__________UUCCAGACUCAAAUGGUGACUCAAAGC .......((((((..((((((((...........)))))))).............((((((((((.......)))))))))).........................))))))....... (-17.48 = -17.76 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:16 2006