
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,367,010 – 8,367,202 |
| Length | 192 |
| Max. P | 0.999979 |

| Location | 8,367,010 – 8,367,123 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.44 |
| Mean single sequence MFE | -38.82 |
| Consensus MFE | -34.16 |
| Energy contribution | -34.08 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8367010 113 + 22224390 UGUUAGUUUCGCUUGUUUUAUCAGCCGUUUUUGCGGCUGCUCUAGGUGCCAGUACCGCUGCUCUGCCGGCGUCUAAAAGUGCAUUGCACUUUUAAGUGUUUUCGGCCUGGG-------AA ..........((.........(((((((....))))))).....((((....))))))..(((.(((((((..(((((((((...)))))))))..)))...))))..)))-------.. ( -38.40) >DroSec_CAF1 32177 113 + 1 UGUUAGUUUCGCUUGUUUUAUCAGCCGUUUUUGCGGCUGCUCUAGGUGCCAGUACCGCUGCUCUGCCGGCGUCUAAAAGUGCAUUGCACUUUUAAGUGUUUUCGGCCUGGU-------AA ..........((.........(((((((....))))))).....((((....))))))((((..(((((((..(((((((((...)))))))))..)))...))))..)))-------). ( -39.00) >DroSim_CAF1 35366 113 + 1 UGUUAGUUUCGCUUGUUUUAUCAGCCGUUUUUGCGGCUGCUCUAGGUGCCAGUACCGCUGCUCUGCCGGCGUCUAAAAGUGCAUUGCACUUUUAAGUGUUUUCGGCCUGGG-------AA ..........((.........(((((((....))))))).....((((....))))))..(((.(((((((..(((((((((...)))))))))..)))...))))..)))-------.. ( -38.40) >DroEre_CAF1 28882 113 + 1 GCUUGGUUUCGCUUGUUUUAUCAGCCGUUUUUGCGGCUUGUUUAGGUGCUAGUACCGCUGCUCUGCCGGCGUCAAAAAGUGCAUUGCACUUUUAAGUGUUUUCGGGCUGGG-------AA ((((((...(((((.........(((((....))(((..((...((((....))))...))...))))))....((((((((...)))))))))))))...))))))....-------.. ( -36.30) >DroYak_CAF1 31111 120 + 1 UGUUAGUUUCGCCUGUUUUAUCAGCCGUUUUUGCGGCUGCUCUAGGUGCCAGUACCGCUGCUCUGCCGGCGUCUGAAAGUGCAUUGCACUUUUAAGUGUUUUCGGCCUGGGAAAUGGGAA ...........((((((((..(((((((....)))))))..(((((.((.((.....)))).).(((((((..(((((((((...)))))))))..)))...)))))))))))))))).. ( -42.00) >consensus UGUUAGUUUCGCUUGUUUUAUCAGCCGUUUUUGCGGCUGCUCUAGGUGCCAGUACCGCUGCUCUGCCGGCGUCUAAAAGUGCAUUGCACUUUUAAGUGUUUUCGGCCUGGG_______AA .........((((((......(((((((....)))))))...))))))((((..((((((......)))))..(((((((((...))))))))).........)..)))).......... (-34.16 = -34.08 + -0.08)

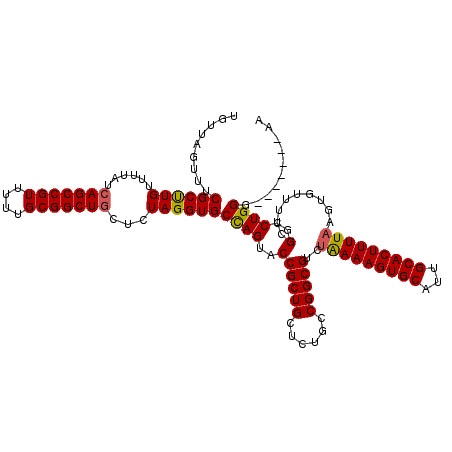
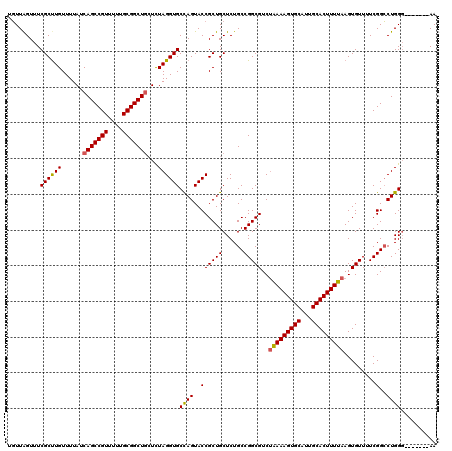
| Location | 8,367,010 – 8,367,123 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.44 |
| Mean single sequence MFE | -30.26 |
| Consensus MFE | -27.26 |
| Energy contribution | -28.26 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.996358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8367010 113 - 22224390 UU-------CCCAGGCCGAAAACACUUAAAAGUGCAAUGCACUUUUAGACGCCGGCAGAGCAGCGGUACUGGCACCUAGAGCAGCCGCAAAAACGGCUGAUAAAACAAGCGAAACUAACA ((-------(((((((((.......((((((((((...))))))))))..(((....).))..)))).)))).........((((((......))))))...........)))....... ( -31.80) >DroSec_CAF1 32177 113 - 1 UU-------ACCAGGCCGAAAACACUUAAAAGUGCAAUGCACUUUUAGACGCCGGCAGAGCAGCGGUACUGGCACCUAGAGCAGCCGCAAAAACGGCUGAUAAAACAAGCGAAACUAACA ..-------.((((((((.......((((((((((...))))))))))..(((....).))..)))).)))).........((((((......))))))..................... ( -31.90) >DroSim_CAF1 35366 113 - 1 UU-------CCCAGGCCGAAAACACUUAAAAGUGCAAUGCACUUUUAGACGCCGGCAGAGCAGCGGUACUGGCACCUAGAGCAGCCGCAAAAACGGCUGAUAAAACAAGCGAAACUAACA ((-------(((((((((.......((((((((((...))))))))))..(((....).))..)))).)))).........((((((......))))))...........)))....... ( -31.80) >DroEre_CAF1 28882 113 - 1 UU-------CCCAGCCCGAAAACACUUAAAAGUGCAAUGCACUUUUUGACGCCGGCAGAGCAGCGGUACUAGCACCUAAACAAGCCGCAAAAACGGCUGAUAAAACAAGCGAAACCAAGC ..-------..(((((...........((((((((...))))))))....(((....).)).(((((................)))))......))))).........((........)) ( -23.69) >DroYak_CAF1 31111 120 - 1 UUCCCAUUUCCCAGGCCGAAAACACUUAAAAGUGCAAUGCACUUUCAGACGCCGGCAGAGCAGCGGUACUGGCACCUAGAGCAGCCGCAAAAACGGCUGAUAAAACAGGCGAAACUAACA (((((.....((((((((..........(((((((...))))))).....(((....).))..)))).)))).........((((((......))))))........)).)))....... ( -32.10) >consensus UU_______CCCAGGCCGAAAACACUUAAAAGUGCAAUGCACUUUUAGACGCCGGCAGAGCAGCGGUACUGGCACCUAGAGCAGCCGCAAAAACGGCUGAUAAAACAAGCGAAACUAACA ..........((((((((.......((((((((((...))))))))))..(((....).))..)))).)))).........((((((......))))))..................... (-27.26 = -28.26 + 1.00)



| Location | 8,367,050 – 8,367,162 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.59 |
| Mean single sequence MFE | -44.46 |
| Consensus MFE | -42.40 |
| Energy contribution | -42.88 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.95 |
| SVM decision value | 5.20 |
| SVM RNA-class probability | 0.999979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8367050 112 + 22224390 UCUAGGUGCCAGUACCGCUGCUCUGCCGGCGUCUAAAAGUGCAUUGCACUUUUAAGUGUUUUCGGCCUGGG-------AAAUCGGAUGGAAAGCGG-GAAAAGACGCCAGAGAAAAGCCC ....((((....))))(((.((((...(((((((((((((((...)))))))))..(((((((.(.((((.-------...)))).).))))))).-.....))))))))))...))).. ( -41.00) >DroSec_CAF1 32217 113 + 1 UCUAGGUGCCAGUACCGCUGCUCUGCCGGCGUCUAAAAGUGCAUUGCACUUUUAAGUGUUUUCGGCCUGGU-------AAAUCGGGUGGGAAGCGGGGAAAAGACCCCAGAGAAAAGCCU ....((((....))))(((.((((((..(((..(((((((((...)))))))))..)))((((.((((((.-------...)))))).))))))((((......))))))))...))).. ( -43.80) >DroSim_CAF1 35406 113 + 1 UCUAGGUGCCAGUACCGCUGCUCUGCCGGCGUCUAAAAGUGCAUUGCACUUUUAAGUGUUUUCGGCCUGGG-------AAAUCGGGUGGAAAGCGGGGAAAAGACGCCAGAGAAAAGCCC ....((((....))))(((.((((...(((((((((((((((...)))))))))..(((((((.((((((.-------...)))))).))))))).......))))))))))...))).. ( -47.90) >DroEre_CAF1 28922 112 + 1 UUUAGGUGCUAGUACCGCUGCUCUGCCGGCGUCAAAAAGUGCAUUGCACUUUUAAGUGUUUUCGGGCUGGG-------AAAUCGGGUGGAAAGCGG-GAAAAGACGCCAGAGAAAAGCCC ....((((....))))(((.((((...((((((.((((((((...))))))))...(((((((.(.((((.-------...)))).).))))))).-.....))))))))))...))).. ( -42.40) >DroYak_CAF1 31151 119 + 1 UCUAGGUGCCAGUACCGCUGCUCUGCCGGCGUCUGAAAGUGCAUUGCACUUUUAAGUGUUUUCGGCCUGGGAAAUGGGAAAUCGGGUGGAAAGCGG-GAAAAGACGCCAGAGAAAAGCCC ....((((....))))(((.((((...(((((((((((((((...))))))))...(((((((.((((((...........)))))).))))))).-....)))))))))))...))).. ( -47.20) >consensus UCUAGGUGCCAGUACCGCUGCUCUGCCGGCGUCUAAAAGUGCAUUGCACUUUUAAGUGUUUUCGGCCUGGG_______AAAUCGGGUGGAAAGCGG_GAAAAGACGCCAGAGAAAAGCCC ....((((....))))(((.((((...(((((((((((((((...)))))))))..(((((((.((((((...........)))))).))))))).......))))))))))...))).. (-42.40 = -42.88 + 0.48)

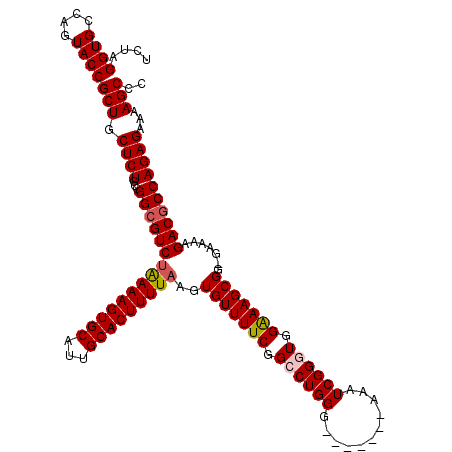
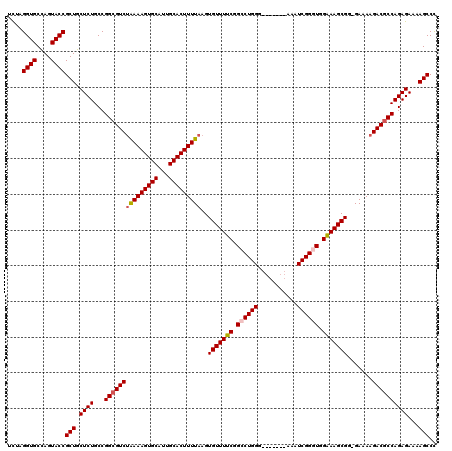
| Location | 8,367,050 – 8,367,162 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.59 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -31.05 |
| Energy contribution | -31.85 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8367050 112 - 22224390 GGGCUUUUCUCUGGCGUCUUUUC-CCGCUUUCCAUCCGAUUU-------CCCAGGCCGAAAACACUUAAAAGUGCAAUGCACUUUUAGACGCCGGCAGAGCAGCGGUACUGGCACCUAGA ..(((.(((((((((((((((((-..((((............-------...)))).)))))....(((((((((...))))))))))))))))).)))).)))(((......))).... ( -35.66) >DroSec_CAF1 32217 113 - 1 AGGCUUUUCUCUGGGGUCUUUUCCCCGCUUCCCACCCGAUUU-------ACCAGGCCGAAAACACUUAAAAGUGCAAUGCACUUUUAGACGCCGGCAGAGCAGCGGUACUGGCACCUAGA ..(((.((((((((.((((((((...((((............-------...)))).)))))....(((((((((...)))))))))))).)))).)))).)))(((......))).... ( -31.36) >DroSim_CAF1 35406 113 - 1 GGGCUUUUCUCUGGCGUCUUUUCCCCGCUUUCCACCCGAUUU-------CCCAGGCCGAAAACACUUAAAAGUGCAAUGCACUUUUAGACGCCGGCAGAGCAGCGGUACUGGCACCUAGA ..(((.((((((((((((........(.((((..((.(....-------..).))..)))).)...(((((((((...))))))))))))))))).)))).)))(((......))).... ( -36.50) >DroEre_CAF1 28922 112 - 1 GGGCUUUUCUCUGGCGUCUUUUC-CCGCUUUCCACCCGAUUU-------CCCAGCCCGAAAACACUUAAAAGUGCAAUGCACUUUUUGACGCCGGCAGAGCAGCGGUACUAGCACCUAAA ..(((.(((((((((((((((((-..(((.............-------...)))..))))).....((((((((...)))))))).)))))))).)))).)))(((......))).... ( -33.59) >DroYak_CAF1 31151 119 - 1 GGGCUUUUCUCUGGCGUCUUUUC-CCGCUUUCCACCCGAUUUCCCAUUUCCCAGGCCGAAAACACUUAAAAGUGCAAUGCACUUUCAGACGCCGGCAGAGCAGCGGUACUGGCACCUAGA ..(((.(((((((((((((....-.............(.((((((........))..)))).).....(((((((...))))))).))))))))).)))).)))(((......))).... ( -33.00) >consensus GGGCUUUUCUCUGGCGUCUUUUC_CCGCUUUCCACCCGAUUU_______CCCAGGCCGAAAACACUUAAAAGUGCAAUGCACUUUUAGACGCCGGCAGAGCAGCGGUACUGGCACCUAGA ..(((.(((((((((((((((((...((((......................)))).)))))....(((((((((...))))))))))))))))).)))).)))(((......))).... (-31.05 = -31.85 + 0.80)



| Location | 8,367,090 – 8,367,202 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.15 |
| Mean single sequence MFE | -29.05 |
| Consensus MFE | -24.24 |
| Energy contribution | -24.68 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8367090 112 + 22224390 GCAUUGCACUUUUAAGUGUUUUCGGCCUGGG-------AAAUCGGAUGGAAAGCGG-GAAAAGACGCCAGAGAAAAGCCCCAUGUGGCACAGUUGCAAAUAGACGAAAAAAAGAAGUAGA ...((((((((((...(((((((.(.((((.-------...)))).).))))))).-.)))))..((((.(...........).)))).....)))))...................... ( -23.10) >DroSec_CAF1 32257 110 + 1 GCAUUGCACUUUUAAGUGUUUUCGGCCUGGU-------AAAUCGGGUGGGAAGCGGGGAAAAGACCCCAGAGAAAAGCCUCAUGUGGCACAGUUGCAAAUAGACGAAAA-A--AUGUAGA ...(((((((((.....((((((.((((((.-------...)))))).))))))((((......))))))))....(((......))).....)))))...........-.--....... ( -33.40) >DroSim_CAF1 35446 111 + 1 GCAUUGCACUUUUAAGUGUUUUCGGCCUGGG-------AAAUCGGGUGGAAAGCGGGGAAAAGACGCCAGAGAAAAGCCCCAUGUGGCACAGUUGCAAAUAGACGAAAAAA--AAGUAGA ...((((((((((...(((((((.((((((.-------...)))))).)))))))...)))))..((((.(...........).)))).....))))).............--....... ( -30.30) >DroEre_CAF1 28962 108 + 1 GCAUUGCACUUUUAAGUGUUUUCGGGCUGGG-------AAAUCGGGUGGAAAGCGG-GAAAAGACGCCAGAGAAAAGCCCCAUGUGGCACAGUUGCACAUACACGAAAA----AGGUAAA ...((((.(((((..((((....(((((...-------...((.((((........-.......)))).))....))))).(((((.((....)))))))))))..)))----)))))). ( -29.46) >DroYak_CAF1 31191 105 + 1 GCAUUGCACUUUUAAGUGUUUUCGGCCUGGGAAAUGGGAAAUCGGGUGGAAAGCGG-GAAAAGACGCCAGAGAAAAGCCCCAUGUGGCACAGUUGCA----------AA----AAGUAGA ...((((((((((...(((((((.((((((...........)))))).))))))).-.)))))..((((.(...........).)))).....))))----------).----....... ( -29.00) >consensus GCAUUGCACUUUUAAGUGUUUUCGGCCUGGG_______AAAUCGGGUGGAAAGCGG_GAAAAGACGCCAGAGAAAAGCCCCAUGUGGCACAGUUGCAAAUAGACGAAAA_A__AAGUAGA ((((((..(((((...(((((((.((((((...........)))))).)))))))...)))))..((((.(...........).)))).))).)))........................ (-24.24 = -24.68 + 0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:11 2006