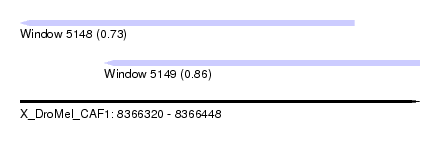
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,366,320 – 8,366,448 |
| Length | 128 |
| Max. P | 0.862491 |

| Location | 8,366,320 – 8,366,427 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.29 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -10.04 |
| Energy contribution | -12.22 |
| Covariance contribution | 2.19 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8366320 107 - 22224390 UUUCAAAUAACUAAAUGUUGGUUCAUUUACCAGAAAGCAUGGUAUGCCAUGCAAAUAGCAAAUCGGAUAUGAUAUAUGUAU---A----------UAGUUUGUUUAAAUAGGGAAAAAAA ((((.......((((((......))))))((.....((((((....))))))....(((((((...((((.......))))---.----------..)))))))......)))))).... ( -21.50) >DroSec_CAF1 31449 114 - 1 UUUCUUAUAACAAAAUGUUGAUUUAUUUACCAGAGAGCAUGGUAUGCCAUGCAAACAGCAAAUCGGAUAUGAAAUAUAUAGUGUAGUG------GCAGUUUGUUGAAAUAGGAAAAAAAA ((((((((((((((.((((.................((((((....))))))..((.(((......(((((...)))))..))).)))------))).))))))...))))))))..... ( -23.10) >DroSim_CAF1 34643 110 - 1 UUUCUUAUAACACAAUGUUGGUUUAUUUACCAGAGAGCAUGGUAUGCCAUGCAAACAGCAAGUCGGAUAUGAAAUAUAUAGUGAAAU----------AUUUGUUGAAAUAGGGAAAAAAA ((((((((.........(((((......)))))...((((((....))))))...((((((((((.(((((...)))))..)))...----------.)))))))..))))))))..... ( -24.80) >DroYak_CAF1 30386 105 - 1 CUGCUAAUAAAUAAAAUUUGGUUUAUUUACCAGAAAACAUGGCAUAUU------AAAGCCAAUCGGAAACGAAAUAUUCAGUGGAGUGGCAGUGGCAGUUUGUU---------AGAAAAA ...(((((((((....((((((......))))))..............------...((((.(((....)))..(((((....)))))....)))).)))))))---------))..... ( -26.20) >consensus UUUCUAAUAACAAAAUGUUGGUUUAUUUACCAGAAAGCAUGGUAUGCCAUGCAAACAGCAAAUCGGAUAUGAAAUAUAUAGUG_AGU________CAGUUUGUUGAAAUAGGGAAAAAAA ((((((((.........(((((......)))))...((((((....))))))..........(((....)))...................................))))))))..... (-10.04 = -12.22 + 2.19)

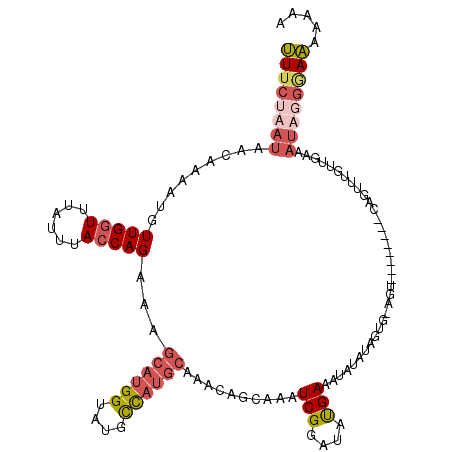
| Location | 8,366,347 – 8,366,448 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.16 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -14.66 |
| Energy contribution | -14.85 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8366347 101 - 22224390 ------------------AAUU-GGUAAUCUUGAUGAGCGUUUCAAAUAACUAAAUGUUGGUUCAUUUACCAGAAAGCAUGGUAUGCCAUGCAAAUAGCAAAUCGGAUAUGAUAUAUGUA ------------------..((-(((((...((((.(((((((.........))))))).).))).)))))))...((((((....)))))).....(((.((((....))))...))). ( -23.80) >DroSec_CAF1 31483 110 - 1 C---------AUACACUCGAUG-GGUAAUCUUGAUAAGCGUUUCUUAUAACAAAAUGUUGAUUUAUUUACCAGAGAGCAUGGUAUGCCAUGCAAACAGCAAAUCGGAUAUGAAAUAUAUA (---------(((...(((((.-(((((....(((.(((((((.........))))))).)))...))))).....((((((....)))))).........))))).))))......... ( -24.20) >DroSim_CAF1 34673 110 - 1 C---------AUACACUCGAUG-GGUAAUCCUGAUGAGCGUUUCUUAUAACACAAUGUUGGUUUAUUUACCAGAGAGCAUGGUAUGCCAUGCAAACAGCAAGUCGGAUAUGAAAUAUAUA (---------(((.(((.....-)))....(((((..(((((......)))....(((((((......))).....((((((....)))))).))))))..))))).))))......... ( -25.90) >DroYak_CAF1 30417 114 - 1 CAUUUGCCGCAUACACUGUAAACGAGGUUUUUAAUGAGCACUGCUAAUAAAUAAAAUUUGGUUUAUUUACCAGAAAACAUGGCAUAUU------AAAGCCAAUCGGAAACGAAAUAUUCA .................((((((.(((((((.....(((...))).......))))))).))))))......(((....((((.....------...)))).(((....)))....))). ( -20.90) >consensus C_________AUACACUCGAUG_GGUAAUCUUGAUGAGCGUUUCUAAUAACAAAAUGUUGGUUUAUUUACCAGAAAGCAUGGUAUGCCAUGCAAACAGCAAAUCGGAUAUGAAAUAUAUA ...........................((((.(((..((..................(((((......)))))...((((((....)))))).....))..)))))))............ (-14.66 = -14.85 + 0.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:07 2006