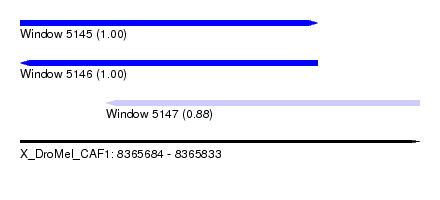
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,365,684 – 8,365,833 |
| Length | 149 |
| Max. P | 0.999802 |

| Location | 8,365,684 – 8,365,795 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 88.38 |
| Mean single sequence MFE | -28.56 |
| Consensus MFE | -23.88 |
| Energy contribution | -24.08 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.998425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8365684 111 + 22224390 UUGUUAUAAUUAAAAUGGAACGCCCCAAAAAGUUGGGGGAGCUGGUAGGAAUCAGCAGGCAUAUAUACUCGUACACAAAUUUGUGCGUUCCAAGUGGAUAAAUACAUACAU- .(((....(((....((((((.((((((....))))))..((((((....))))))..............(((((......)))))))))))....)))....))).....- ( -30.30) >DroSec_CAF1 30900 108 + 1 UUGUUAUAAUUAAAAUGGAACGCCCCAAAAAGUUGGGGGAGCUGGUAGGAAUCAGCAG---UAUAUACUCGUAUACAAAUUUGUGCGUUCCAAGUGGAUAAAUACAUACAU- .(((....(((....(((((((((((((....)))))...((((((....)))))).(---(((((....))))))........))))))))....)))....))).....- ( -31.00) >DroSim_CAF1 34079 108 + 1 UUGUUAUAAUUAAAAUGGAACGCCCCAAAAAGUUGGGGGAGCUGUUAGGAAUCAGCAG---CAUAUACUCGUAUACAAAUUUGUGCGUUCCAAGUGGAUA-AUACAUACAUA .((((((.....(..((((((.((((((....))))))..((((((.......)))))---)........(((((......)))))))))))..)..)))-)))........ ( -28.20) >DroEre_CAF1 28021 101 + 1 UUGUUAUAAUUAAAAUGGAACGCCCCAAAAAGUUGGGGGAACUGGUAGGAAUCAGCAG---CAUAUAC------AAAAAUGUGUGCUUUCCAAGUGGAUAAAUACAUACA-- .(((....(((....(((((..((((((....))))))...(((((....))))).((---((((((.------.....)))))))))))))....)))....)))....-- ( -24.30) >DroYak_CAF1 29719 103 + 1 UUGUUAUAAUUAAAAUGGAACGCCCCAGAAAGUUGGGCAAACUGGUAGGAAUCAGUGG---CAUAUAC------AAAAAUGUGUGCGUUCCAAGUGGAUAACUACAUAUACA ...............(((((((((((((....)))))...((((((....)))))).(---(((((..------....)))))))))))))).((((....))))....... ( -29.00) >consensus UUGUUAUAAUUAAAAUGGAACGCCCCAAAAAGUUGGGGGAGCUGGUAGGAAUCAGCAG___CAUAUACUCGUA_ACAAAUUUGUGCGUUCCAAGUGGAUAAAUACAUACAU_ .(((....(((....(((((((((((((....)))))...((((((....))))))............................))))))))....)))....)))...... (-23.88 = -24.08 + 0.20)



| Location | 8,365,684 – 8,365,795 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 88.38 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -23.18 |
| Energy contribution | -23.58 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.88 |
| SVM decision value | 4.11 |
| SVM RNA-class probability | 0.999802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8365684 111 - 22224390 -AUGUAUGUAUUUAUCCACUUGGAACGCACAAAUUUGUGUACGAGUAUAUAUGCCUGCUGAUUCCUACCAGCUCCCCCAACUUUUUGGGGCGUUCCAUUUUAAUUAUAACAA -.(((.((((.(((......((((((((.......((((((....)))))).....((((........))))...(((((....)))))))))))))...))).))))))). ( -29.00) >DroSec_CAF1 30900 108 - 1 -AUGUAUGUAUUUAUCCACUUGGAACGCACAAAUUUGUAUACGAGUAUAUA---CUGCUGAUUCCUACCAGCUCCCCCAACUUUUUGGGGCGUUCCAUUUUAAUUAUAACAA -.(((.((((.(((......((((((((.......((((((....))))))---..((((........))))...(((((....)))))))))))))...))).))))))). ( -28.90) >DroSim_CAF1 34079 108 - 1 UAUGUAUGUAU-UAUCCACUUGGAACGCACAAAUUUGUAUACGAGUAUAUG---CUGCUGAUUCCUAACAGCUCCCCCAACUUUUUGGGGCGUUCCAUUUUAAUUAUAACAA ..(((.((((.-........((((((((........(((((......))))---).((((........))))...(((((....))))))))))))).......))))))). ( -26.99) >DroEre_CAF1 28021 101 - 1 --UGUAUGUAUUUAUCCACUUGGAAAGCACACAUUUUU------GUAUAUG---CUGCUGAUUCCUACCAGUUCCCCCAACUUUUUGGGGCGUUCCAUUUUAAUUAUAACAA --(((.((((.(((......(((((((((.(((....)------))...))---))((((........))))..((((((....))))))..)))))...))).))))))). ( -21.10) >DroYak_CAF1 29719 103 - 1 UGUAUAUGUAGUUAUCCACUUGGAACGCACACAUUUUU------GUAUAUG---CCACUGAUUCCUACCAGUUUGCCCAACUUUCUGGGGCGUUCCAUUUUAAUUAUAACAA ......((((((((......((((((((..........------.......---..((((........))))...((((......))))))))))))...)))))))).... ( -25.90) >consensus _AUGUAUGUAUUUAUCCACUUGGAACGCACAAAUUUGU_UACGAGUAUAUG___CUGCUGAUUCCUACCAGCUCCCCCAACUUUUUGGGGCGUUCCAUUUUAAUUAUAACAA ..(((.((((.(((......((((((((............................((((........))))...(((((....)))))))))))))...))).))))))). (-23.18 = -23.58 + 0.40)

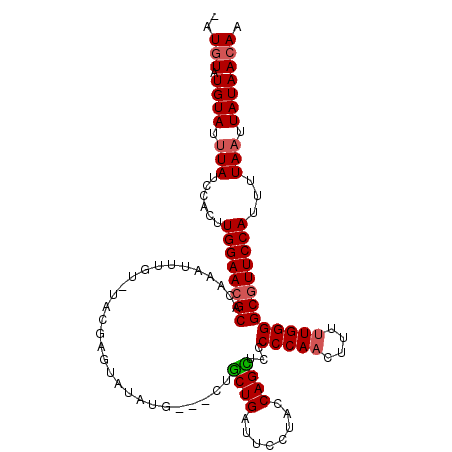
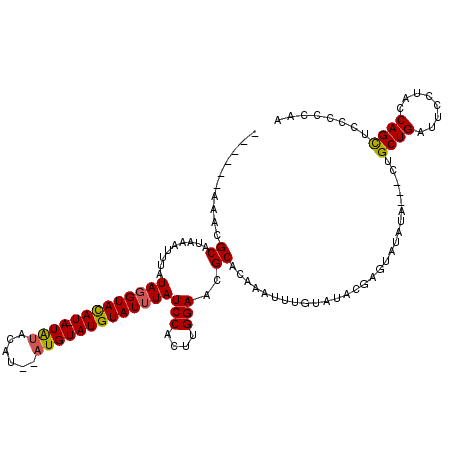
| Location | 8,365,716 – 8,365,833 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.55 |
| Mean single sequence MFE | -22.73 |
| Consensus MFE | -14.56 |
| Energy contribution | -14.50 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8365716 117 - 22224390 GCACAAAAACGCAUAAAUUUAUAGGUACAU-AUAUACAU--AUGUAUGUAUUUAUCCACUUGGAACGCACAAAUUUGUGUACGAGUAUAUAUGCCUGCUGAUUCCUACCAGCUCCCCCAA ..........(((((.....((((((((((-((((....--))))))))))))))..(((((..(((((......))))).)))))...)))))..((((........))))........ ( -29.50) >DroSec_CAF1 30932 113 - 1 NNNN-NAAACGCAUAAAUUUAUAGGUACAU-AUAUGCAU--AUGUAUGUAUUUAUCCACUUGGAACGCACAAAUUUGUAUACGAGUAUAUA---CUGCUGAUUCCUACCAGCUCCCCCAA ....-.....((.........(((((((((-((((....--)))))))))))))(((....)))..)).......((((((....))))))---..((((........))))........ ( -22.50) >DroSim_CAF1 34111 114 - 1 GCAC-AAAACGCAUAAAUUUAUAGGUACAU-AUAUGCAUAUAUGUAUGUAU-UAUCCACUUGGAACGCACAAAUUUGUAUACGAGUAUAUG---CUGCUGAUUCCUAACAGCUCCCCCAA ....-.....(((((........(((((((-(((((....)))))))))))-)....(((((....(((......)))...))))).))))---).((((........))))........ ( -23.70) >DroEre_CAF1 28053 96 - 1 ---------CGCAUAAAUUUAUAGGUAUAUAAUGCA------UGUAUGUAUUUAUCCACUUGGAAAGCACACAUUUUU------GUAUAUG---CUGCUGAUUCCUACCAGUUCCCCCAA ---------..............((((((((....(------(((.(((.....(((....)))..))).))))....------.))))))---))((((........))))........ ( -15.20) >consensus ______AAACGCAUAAAUUUAUAGGUACAU_AUAUACAU__AUGUAUGUAUUUAUCCACUUGGAACGCACAAAUUUGUAUACGAGUAUAUA___CUGCUGAUUCCUACCAGCUCCCCCAA ..........((.........(((((((((.((((......)))))))))))))(((....)))..))............................((((........))))........ (-14.56 = -14.50 + -0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:05 2006