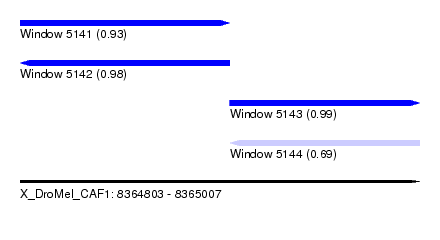
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,364,803 – 8,365,007 |
| Length | 204 |
| Max. P | 0.992629 |

| Location | 8,364,803 – 8,364,910 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 82.58 |
| Mean single sequence MFE | -27.17 |
| Consensus MFE | -20.12 |
| Energy contribution | -21.12 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8364803 107 + 22224390 ---UUUUAUUUCAAGCACAGUGUGAUGAUAAUUUUCGGCCAUUCUUGACUUUGAAAAAAUCAAGGCCAAGGUGAUCGGAGAAUAUUUCCAUGUUGCAUGUGAAAG-ACUAU ---.....(((((....(((..((..((((.((((((..(((.((((.((((((.....)))))).)))))))..)))))).))))..))..)))....))))).-..... ( -30.30) >DroSec_CAF1 29985 110 + 1 UUCUUUUUUUUCAAGCACAGUAGGAAGAUAAUAUUCGGCGAUCCUUGACUUCGAAAAAAGCAAGGCCAAGGUGAUCGGAGAAUACUUCCAAGUUGCAGGUGAAAG-ACUAU ......(((((((.(((.(.(.(((((....(((((..(((((((((.(((......)))))))((....)))))))..)))))))))).).))))...))))))-).... ( -27.30) >DroSim_CAF1 33181 110 + 1 UUCUUUUUUUUCAAGCAUAGUAGAAAGAUAAUAUUCGGCGAUCCUUGACUUUGAAUAAAUCAAGGCCAAGGUGAUCGGAGAAUACUUCCAAGUUGCAGGUGAAAG-ACUAU ......(((((((.(((.....(((.(....(((((..(((((((((.((((((.....)))))).)))))..))))..))))))))).....)))...))))))-).... ( -25.70) >DroEre_CAF1 27094 107 + 1 ----ACGUUUUCAAGCACAAUAUAAAGAUAAUUUUUGACAAUCCUUGACUUUGAAAAAAUCAAGGCCAAGGUGUUCAGAAAAUACGUGGAAGUUGCAAGUGAAAACUCCAU ----..(((((((.(((..............(((((((....(((((.((((((.....)))))).)))))...)))))))..((......)))))...)))))))..... ( -25.40) >consensus ___UUUUUUUUCAAGCACAGUAGAAAGAUAAUAUUCGGCGAUCCUUGACUUUGAAAAAAUCAAGGCCAAGGUGAUCGGAGAAUACUUCCAAGUUGCAGGUGAAAG_ACUAU ........(((((.(((.....(((((.((.(((((((....(((((.((((((.....)))))).)))))...))))))).)))))))....)))...)))))....... (-20.12 = -21.12 + 1.00)


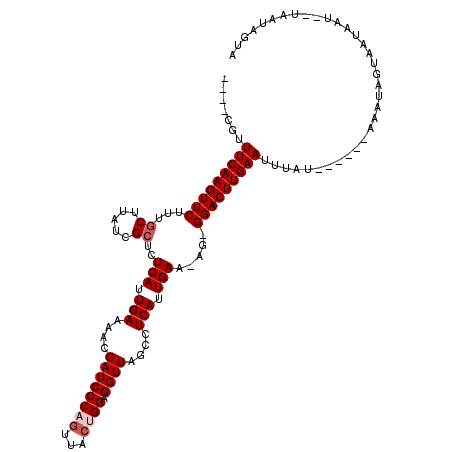
| Location | 8,364,803 – 8,364,910 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 82.58 |
| Mean single sequence MFE | -25.61 |
| Consensus MFE | -18.51 |
| Energy contribution | -19.82 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8364803 107 - 22224390 AUAGU-CUUUCACAUGCAACAUGGAAAUAUUCUCCGAUCACCUUGGCCUUGAUUUUUUCAAAGUCAAGAAUGGCCGAAAAUUAUCAUCACACUGUGCUUGAAAUAAAA--- .....-.(((((...(((.((.(((.......)))(((....(((((((((((((.....)))))))....)))))).....))).......))))).))))).....--- ( -20.20) >DroSec_CAF1 29985 110 - 1 AUAGU-CUUUCACCUGCAACUUGGAAGUAUUCUCCGAUCACCUUGGCCUUGCUUUUUUCGAAGUCAAGGAUCGCCGAAUAUUAUCUUCCUACUGUGCUUGAAAAAAAAGAA .....-.(((((...(((....((((((((((..((((..(((((((.(((.......))).)))))))))))..)))))....))))).....))).)))))........ ( -28.00) >DroSim_CAF1 33181 110 - 1 AUAGU-CUUUCACCUGCAACUUGGAAGUAUUCUCCGAUCACCUUGGCCUUGAUUUAUUCAAAGUCAAGGAUCGCCGAAUAUUAUCUUUCUACUAUGCUUGAAAAAAAAGAA .....-.(((((...(((...(((((((((((..((((..(((((((.((((.....)))).)))))))))))..)))))).....)))))...))).)))))........ ( -28.20) >DroEre_CAF1 27094 107 - 1 AUGGAGUUUUCACUUGCAACUUCCACGUAUUUUCUGAACACCUUGGCCUUGAUUUUUUCAAAGUCAAGGAUUGUCAAAAAUUAUCUUUAUAUUGUGCUUGAAAACGU---- .....(((((((..(((((.......(((.(((.(((...(((((((.((((.....)))).)))))))....))).))).))).......)))))..)))))))..---- ( -26.04) >consensus AUAGU_CUUUCACCUGCAACUUGGAAGUAUUCUCCGAUCACCUUGGCCUUGAUUUUUUCAAAGUCAAGGAUCGCCGAAAAUUAUCUUCAUACUGUGCUUGAAAAAAAA___ .......(((((...(((....((((((((((..((((..(((((((.((((.....)))).)))))))))))..)))))))....))).....))).)))))........ (-18.51 = -19.82 + 1.31)



| Location | 8,364,910 – 8,365,007 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.98 |
| Mean single sequence MFE | -26.34 |
| Consensus MFE | -19.61 |
| Energy contribution | -19.37 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8364910 97 + 22224390 UACUAUUA------------UUU------AUAAAUUCAAGUCCCCU-UUCAAUGAGGCUAACCUCCAGUAACUGGGAUCGUUUUCAAUGAGAGCGAUAAGAAAAGAACUUGAAACG---- ........------------...------.....(((((((.(((.-......((((....))))........)))(((((((((...))))))))).........)))))))...---- ( -23.26) >DroSec_CAF1 30095 114 + 1 UACUAUUAGUAUUAUUACUAUGUAACUAUAUAAAUUCAAGUCC-CU-UUCAAUGAGGCUAACCUCCAGUAACUGGGAUCGUUUUCAAUGAGAGCGAUAAGCGAGGAACUUGAAACG---- (((...(((((....))))).)))..........(((((((.(-((-(.(...((((....))))...........(((((((((...)))))))))..).)))).)))))))...---- ( -29.50) >DroSim_CAF1 33291 108 + 1 UACUAUUAGUAUUAUUACUAUGU------AUAAAUUCAAGUCC-CU-UUCAAUGAGGCUAACCUCCAGUAACUGGGAUCGUUUUCAAUGAGAGCGAUAAGCGAGGAACUUGAAACG---- (((...(((((....))))).))------)....(((((((.(-((-(.(...((((....))))...........(((((((((...)))))))))..).)))).)))))))...---- ( -29.30) >DroEre_CAF1 27201 107 + 1 UUCCAACA--AUGACUAAUAUUU------AUAAAUUCAAGUCC-CUUUUCAAUGAGGAUAACCUCCAGUAACGGGGAUCGUUUUCAAUGAAAGCGAUAAGCAAAGAACUUGAAACG---- ........--.............------.....(((((((((-((.......((((....)))).......))))(((((((((...))))))))).........)))))))...---- ( -24.24) >DroYak_CAF1 28980 103 + 1 -----ACU--AUUAC--AUAUUU------AUACAUUCAAGUCC-CU-UUCAAUGAGGAUAACCUCCAUUCAUGGGGAUCGUUUUCAAUGAAAGCGAUAAGCAAAGAACUUGAAACGCUUA -----...--.....--......------.....(((((((((-((-...(((((((....))).))))...))))(((((((((...))))))))).........)))))))....... ( -25.40) >consensus UACUAUUA__AUUACUAAUAUUU______AUAAAUUCAAGUCC_CU_UUCAAUGAGGCUAACCUCCAGUAACUGGGAUCGUUUUCAAUGAGAGCGAUAAGCAAAGAACUUGAAACG____ ..................................(((((((((.(........((((....))))........)))(((((((((...))))))))).........)))))))....... (-19.61 = -19.37 + -0.24)

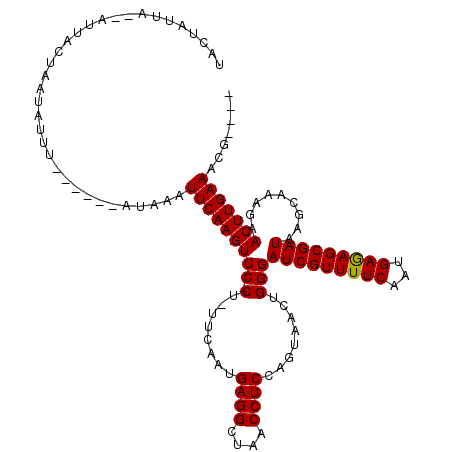

| Location | 8,364,910 – 8,365,007 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.98 |
| Mean single sequence MFE | -24.68 |
| Consensus MFE | -17.10 |
| Energy contribution | -17.90 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8364910 97 - 22224390 ----CGUUUCAAGUUCUUUUCUUAUCGCUCUCAUUGAAAACGAUCCCAGUUACUGGAGGUUAGCCUCAUUGAA-AGGGGACUUGAAUUUAU------AAA------------UAAUAGUA ----...(((((((((((((..........(((.(((....((((((((...)))).))))....))).))))-)))))))))))).....------...------------........ ( -21.50) >DroSec_CAF1 30095 114 - 1 ----CGUUUCAAGUUCCUCGCUUAUCGCUCUCAUUGAAAACGAUCCCAGUUACUGGAGGUUAGCCUCAUUGAA-AG-GGACUUGAAUUUAUAUAGUUACAUAGUAAUAAUACUAAUAGUA ----...(((((((((((.((.....))..(((.(((....((((((((...)))).))))....))).))).-))-)))))))))..............(((((....)))))...... ( -27.30) >DroSim_CAF1 33291 108 - 1 ----CGUUUCAAGUUCCUCGCUUAUCGCUCUCAUUGAAAACGAUCCCAGUUACUGGAGGUUAGCCUCAUUGAA-AG-GGACUUGAAUUUAU------ACAUAGUAAUAAUACUAAUAGUA ----...(((((((((((.((.....))..(((.(((....((((((((...)))).))))....))).))).-))-))))))))).....------...(((((....)))))...... ( -27.30) >DroEre_CAF1 27201 107 - 1 ----CGUUUCAAGUUCUUUGCUUAUCGCUUUCAUUGAAAACGAUCCCCGUUACUGGAGGUUAUCCUCAUUGAAAAG-GGACUUGAAUUUAU------AAAUAUUAGUCAU--UGUUGGAA ----...((((((((((((((.....)).((((.(((..(..(((.(((....))).)))..)..))).)))))))-))))))))).....------.............--........ ( -22.90) >DroYak_CAF1 28980 103 - 1 UAAGCGUUUCAAGUUCUUUGCUUAUCGCUUUCAUUGAAAACGAUCCCCAUGAAUGGAGGUUAUCCUCAUUGAA-AG-GGACUUGAAUGUAU------AAAUAU--GUAAU--AGU----- ...(((((.((((((((((((.....))..(((.(((..(..(((.(((....))).)))..)..))).))))-))-))))))))))))..------......--.....--...----- ( -24.40) >consensus ____CGUUUCAAGUUCUUUGCUUAUCGCUCUCAUUGAAAACGAUCCCAGUUACUGGAGGUUAGCCUCAUUGAA_AG_GGACUUGAAUUUAU______AAAUAGUAAUAAU__UAAUAGUA .......(((((((((...((.....))..(((.(((....((((((((...)))).))))....))).))).....))))))))).................................. (-17.10 = -17.90 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:02 2006