
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,363,861 – 8,364,052 |
| Length | 191 |
| Max. P | 0.906771 |

| Location | 8,363,861 – 8,363,981 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.89 |
| Mean single sequence MFE | -28.44 |
| Consensus MFE | -25.98 |
| Energy contribution | -25.70 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.844982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8363861 120 - 22224390 UAGGAACGUGGAAAGUACAUGUGAAAAUGCGAGAAGAGCGCCAAAGUGGCGACGAACGAGGGAGAGGGCAAAAAACGUAAAAAGAGCUCAAAAAGUGAUCUCUUGAAGCUGCCCCACCUU .....(((((.......)))))...............(((((.....)))).)....((((..(.(((((......((...(((((.(((.....))).)))))...)))))))).)))) ( -29.10) >DroSec_CAF1 29029 119 - 1 UAGGAACGUGGAAAAUACAUGUGAAAAUACGAGAAGAGCGCCAAAGUGGCGACGAACUAUGGAGAGGGCGAAAAACGUAAAAAGAGCUCAA-AAGUGAUCUCUUGAAGCUGCCCCACCUU .(((..(((((......(.(((....))).)......(((((.....)))).)...)))))..(.(((((......((...(((((.(((.-...))).)))))...)))))))).))). ( -27.60) >DroSim_CAF1 32238 119 - 1 UAGGAACGUGGAAAAUACAUGUGAAAAUACGAGAAGAGCGCCAAAGUGGCGACGAACUAUGGAGAGGGCGAAAAACGUAAAAAGAGCUCAA-AAGUGAUCUCUUGAAGCUGCCCCACCUU .(((..(((((......(.(((....))).)......(((((.....)))).)...)))))..(.(((((......((...(((((.(((.-...))).)))))...)))))))).))). ( -27.60) >DroEre_CAF1 26203 117 - 1 UUUGAACAUGGAAAAUACAUGUGAAAAUGCGAGAAGAGCGCCAAAGUGGCGACGAAUGAGGGAGAAGGCAA---AUGUAAAAAGAGCUCAAAAAGUGAUCUCUUAAAGCUGCCCCACCUU (((..(((((.......)))))..)))..........(((((.....)))).)....((((..(..((((.---..((...(((((.(((.....))).)))))...)))))))..)))) ( -26.10) >DroYak_CAF1 27959 117 - 1 UAUGAACAUGGAAAAUACAUGUGAAAAUGCGAGAAGAGCGCCAAAGUGGCGACGAGCGAGGGAGAGGGCGA---AUGUGAAAAGAGCUCAAAAAGUGAUCUCUUCAAGCUGCCCCACCUU .....(((((.......))))).....(((..(.....((((.....)))).)..)))((((.(.(((((.---...(((..((((.(((.....))).)))))))...)))))).)))) ( -31.80) >consensus UAGGAACGUGGAAAAUACAUGUGAAAAUGCGAGAAGAGCGCCAAAGUGGCGACGAACGAGGGAGAGGGCGAAAAACGUAAAAAGAGCUCAAAAAGUGAUCUCUUGAAGCUGCCCCACCUU .....(((((.......)))))...............(((((.....)))).).......((.(.(((((......((...(((((.(((.....))).)))))...)))))))).)).. (-25.98 = -25.70 + -0.28)

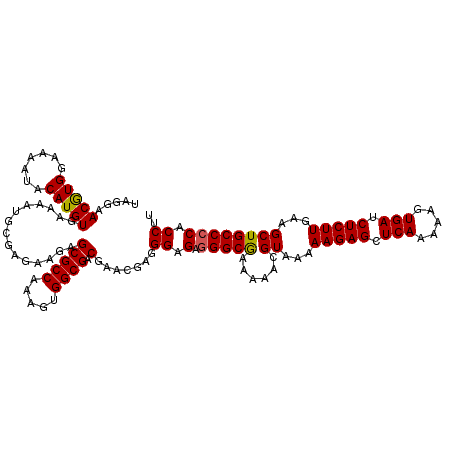
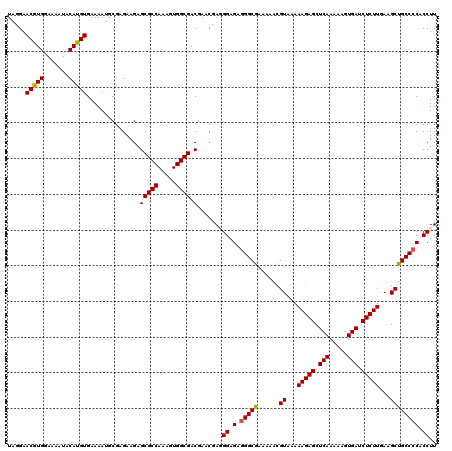
| Location | 8,363,901 – 8,364,020 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -18.74 |
| Energy contribution | -18.34 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8363901 119 - 22224390 CUUGAGGUAUGAAAUCGAAAGAAUUUAGAGAGAAAUCGCUAGGAACGUGGAAAGUACAUGUGAAAAUGCGAGAAGAGCGCCAAAGUGGCGACGAACGAGGGAGAGGGCAAAAAACGUAA ((((.....((((.((....)).))))........((((.....(((((.......)))))......)))).....(((((.....)))).)...)))).................... ( -24.00) >DroSec_CAF1 29068 119 - 1 CUUGAGGUAUCAAAUCAAAAGAAUUUAGAGAGAAAUCGCUAGGAACGUGGAAAAUACAUGUGAAAAUACGAGAAGAGCGCCAAAGUGGCGACGAACUAUGGAGAGGGCGAAAAACGUAA .((((....)))).((.......(((((.((....)).))))).(((((.......)))))))...((((.......((((.....)))).((..((......))..)).....)))). ( -23.90) >DroSim_CAF1 32277 119 - 1 CUUGAGGUAUCAAAUCAAAAGAAUUUAGAGAGAAAUCGCUAGGAACGUGGAAAAUACAUGUGAAAAUACGAGAAGAGCGCCAAAGUGGCGACGAACUAUGGAGAGGGCGAAAAACGUAA .((((....)))).((.......(((((.((....)).))))).(((((.......)))))))...((((.......((((.....)))).((..((......))..)).....)))). ( -23.90) >DroEre_CAF1 26243 116 - 1 CUUGAGGUAUUAAAACGAAAGAAUUUAGAGAGAAAUCGCUUUGAACAUGGAAAAUACAUGUGAAAAUGCGAGAAGAGCGCCAAAGUGGCGACGAAUGAGGGAGAAGGCAA---AUGUAA (((............(....).((((.........(((((((..(((((.......)))))..))).)))).....(((((.....)))).))))))))...........---...... ( -20.40) >DroYak_CAF1 27999 116 - 1 CUUGAGGUAUCAAAUCGAAAGAAUUUAGAGAGAAAUCGCUAUGAACAUGGAAAAUACAUGUGAAAAUGCGAGAAGAGCGCCAAAGUGGCGACGAGCGAGGGAGAGGGCGA---AUGUGA .((((....)))).((....)).............(((((....(((((.......))))).....(((..(.....((((.....)))).)..)))........)))))---...... ( -23.80) >consensus CUUGAGGUAUCAAAUCGAAAGAAUUUAGAGAGAAAUCGCUAGGAACGUGGAAAAUACAUGUGAAAAUGCGAGAAGAGCGCCAAAGUGGCGACGAACGAGGGAGAGGGCGAAAAACGUAA (((..(((.....)))..)))..(((((.((....)).))))).(((((.......))))).....((((......(((((.....)))).)(..(........)..)......)))). (-18.74 = -18.34 + -0.40)

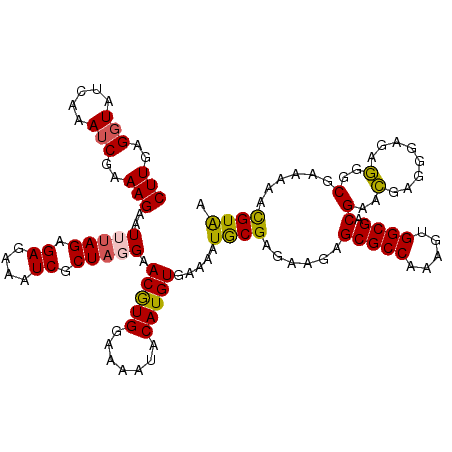
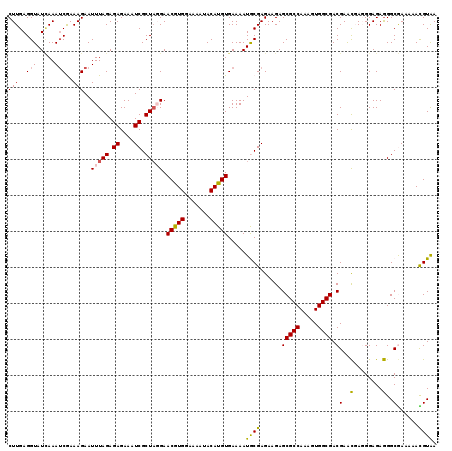
| Location | 8,363,941 – 8,364,052 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 94.19 |
| Mean single sequence MFE | -20.82 |
| Consensus MFE | -16.96 |
| Energy contribution | -17.28 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8363941 111 - 22224390 AA--AUUGUAAAUGGAAAACCCGUUAUCUCUGUACUUGAGGUAUGAAAUCGAAAGAAUUUAGAGAGAAAUCGCUAGGAACGUGGAAAGUACAUGUGAAAAUGCGAGAAGAGCG ..--......(((((.....))))).(((((((((.....))))(((.((....)).)))..)))))..((((.....(((((.......)))))......))))........ ( -23.80) >DroSec_CAF1 29108 111 - 1 AA--AAUGUAAUUGGAAAACCCGUUAUCUCAGUACUUGAGGUAUCAAAUCAAAAGAAUUUAGAGAGAAAUCGCUAGGAACGUGGAAAAUACAUGUGAAAAUACGAGAAGAGCG ..--.........((.....))(((.((((.(((.((((....)))).((.......(((((.((....)).))))).(((((.......)))))))...)))))))..))). ( -18.20) >DroSim_CAF1 32317 111 - 1 AA--AAUGUAAUUGGAAAACCCGUUAUCUCAGUACUUGAGGUAUCAAAUCAAAAGAAUUUAGAGAGAAAUCGCUAGGAACGUGGAAAAUACAUGUGAAAAUACGAGAAGAGCG ..--.........((.....))(((.((((.(((.((((....)))).((.......(((((.((....)).))))).(((((.......)))))))...)))))))..))). ( -18.20) >DroEre_CAF1 26280 113 - 1 AAAAAAUGUAAUUGGAAAACCCGUUAUCUCAGUACUUGAGGUAUUAAAACGAAAGAAUUUAGAGAGAAAUCGCUUUGAACAUGGAAAAUACAUGUGAAAAUGCGAGAAGAGCG .............((.....))(((.(((((((((.....)))))....(....)......))))....(((((((..(((((.......)))))..))).))))....))). ( -22.40) >DroYak_CAF1 28036 111 - 1 AA--AUUGUAAUUGGAAAACCCGUUAUCUCAGUACUUGAGGUAUCAAAUCGAAAGAAUUUAGAGAGAAAUCGCUAUGAACAUGGAAAAUACAUGUGAAAAUGCGAGAAGAGCG ..--.........((.....))(((.((((.....((((....)))).((....)).....))))....((((.....(((((.......)))))......))))....))). ( -21.50) >consensus AA__AAUGUAAUUGGAAAACCCGUUAUCUCAGUACUUGAGGUAUCAAAUCGAAAGAAUUUAGAGAGAAAUCGCUAGGAACGUGGAAAAUACAUGUGAAAAUGCGAGAAGAGCG .............((.....))(((.((((.((((((..(((.....)))..)))..(((((.((....)).))))).(((((.......))))).....)))))))..))). (-16.96 = -17.28 + 0.32)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:59 2006