
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,362,630 – 8,362,790 |
| Length | 160 |
| Max. P | 0.988204 |

| Location | 8,362,630 – 8,362,750 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -31.34 |
| Consensus MFE | -27.90 |
| Energy contribution | -27.94 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.957176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
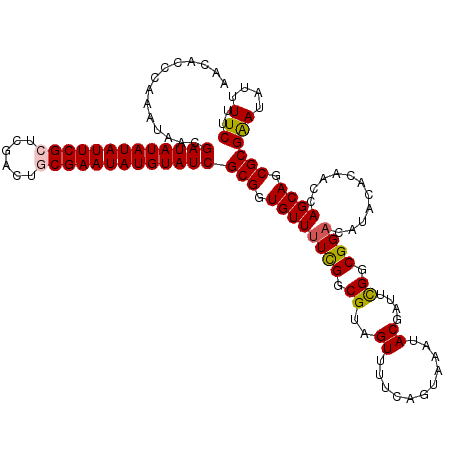
>X_DroMel_CAF1 8362630 120 - 22224390 AACACCCAAAUAACGAUAUAUAUUCGCUCGGCUGCGAAUAUGUAUCGCGGUGUUUUCGGCGUAGUUUUCAGUAAAUACGAUUUGGCGGACAUACACAACCAGCAGCGCGGAUAUUUUCUU .....((.......(((((((((((((......)))))))))))))(((.((((....(.(((((((((((..........)))).)))).))).)....)))).))))).......... ( -34.30) >DroSec_CAF1 27803 120 - 1 AACACCCAAAUAACGAUAUAUAUUCGCUCGACUGCGAAUAUGUAUCGCGGUGUUUUCGGCGUAGUUUUCAGUAAAUACGAUUCGGCGGACAUACAGAACCAGCAGCGCGAAUAUUUUCUU ((((((.......((((((((((((((......)))))))))))))).))))))((((.(((.((((((.(((....((......))....))).)))..))).)))))))......... ( -34.60) >DroSim_CAF1 31000 120 - 1 AACACCCAAAGAACGAUAUAUAUUCGCUCGACUGCGAAUAUGUAUCGCGGUGUUUUCGGCGUAGUUUUCAGUAAAUACGUUUCGGCGGACAUACACAACCAGCAGCGCGAAUAUUUUCUU ........(((((.(((((((((((((......)))))))))))))(((.((((...((((((.((......)).))))))..((..(.......)..)))))).))).......))))) ( -34.00) >DroEre_CAF1 25033 119 - 1 AACAUCCAAAUAACGAUAUAUAUUCGCACGACUACGAAUAUGUAUCGCGGUGUU-UUGGCGUAGUUUUCAGUAAAUACGAUUCGGCGGACAUACACAACCAGCAGCGCGAAUAUUUUCUU ..............((((((((((((........))))))))))))(((.((((-(((..((((((..(.((....))......)..))).))).)))..)))).)))(((....))).. ( -26.60) >DroYak_CAF1 26762 120 - 1 AACAUCCAAAUAACGAUAUAUAUUCGCUCGACUUCGAAUAUGUAUCGCGGUGUUUUUGGCGUAGUUUUCAGCAAAUACGAUUCGGCGGACAUACACAACCAGCAGCGCGAAUAUUUUCUU ..............((((((((((((........))))))))))))(((.((((.(((..((((((....((..(......)..)).))).))).)))..)))).)))(((....))).. ( -27.20) >consensus AACACCCAAAUAACGAUAUAUAUUCGCUCGACUGCGAAUAUGUAUCGCGGUGUUUUCGGCGUAGUUUUCAGUAAAUACGAUUCGGCGGACAUACACAACCAGCAGCGCGAAUAUUUUCUU ..............(((((((((((((......)))))))))))))(((.((((((((.((..((...........))....)).))))...........)))).)))(((....))).. (-27.90 = -27.94 + 0.04)

| Location | 8,362,670 – 8,362,790 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.91 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -20.80 |
| Energy contribution | -20.84 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8362670 120 + 22224390 UCGUAUUUACUGAAAACUACGCCGAAAACACCGCGAUACAUAUUCGCAGCCGAGCGAAUAUAUAUCGUUAUUUGGGUGUUUAAGCACUAACUUUAUUUAUUUUUUCCUAAAAAUUCCGCG ...................(((.(((((((((((((((.((((((((......)))))))).))))))......))))))).................((((((....)))))))).))) ( -27.80) >DroSec_CAF1 27843 119 + 1 UCGUAUUUACUGAAAACUACGCCGAAAACACCGCGAUACAUAUUCGCAGUCGAGCGAAUAUAUAUCGUUAUUUGGGUGUUUAAGCACUAACUUUAUUUAUUUUUUCCUAAA-AUUUCGCG ...................(((.((((.....((((((.((((((((......)))))))).))))))......(((((....))))).......................-.))))))) ( -26.20) >DroSim_CAF1 31040 119 + 1 ACGUAUUUACUGAAAACUACGCCGAAAACACCGCGAUACAUAUUCGCAGUCGAGCGAAUAUAUAUCGUUCUUUGGGUGUUUAAUCACUAACUUUAUUUAUUUUUUCCUACA-AUUUCGCG ......(((.(((.....(((((.(((.....((((((.((((((((......)))))))).))))))..))).)))))....))).))).....................-........ ( -25.40) >DroEre_CAF1 25073 119 + 1 UCGUAUUUACUGAAAACUACGCCAA-AACACCGCGAUACAUAUUCGUAGUCGUGCGAAUAUAUAUCGUUAUUUGGAUGUUUAAGCAUUUAGUCUAUUUAUUUUUUUCUAACAAUUUCGCG ...........((((.....((..(-((((((((((((.(((((((((....))))))))).)))))).....)).)))))..))..((((...............))))...))))... ( -23.06) >DroYak_CAF1 26802 120 + 1 UCGUAUUUGCUGAAAACUACGCCAAAAACACCGCGAUACAUAUUCGAAGUCGAGCGAAUAUAUAUCGUUAUUUGGAUGUUUAAGCACUUACUCUAUUUAUUUUUUCCUUAAAAUUUUGCG ..(((..((((..((((....(((((......((((((.(((((((........))))))).))))))..)))))..)))).))))..)))............................. ( -25.90) >consensus UCGUAUUUACUGAAAACUACGCCGAAAACACCGCGAUACAUAUUCGCAGUCGAGCGAAUAUAUAUCGUUAUUUGGGUGUUUAAGCACUAACUUUAUUUAUUUUUUCCUAAAAAUUUCGCG ...................(((.(((......((((((.((((((((......)))))))).))))))......(((((....))))).........................))).))) (-20.80 = -20.84 + 0.04)


| Location | 8,362,670 – 8,362,790 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.91 |
| Mean single sequence MFE | -28.98 |
| Consensus MFE | -23.84 |
| Energy contribution | -24.36 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8362670 120 - 22224390 CGCGGAAUUUUUAGGAAAAAAUAAAUAAAGUUAGUGCUUAAACACCCAAAUAACGAUAUAUAUUCGCUCGGCUGCGAAUAUGUAUCGCGGUGUUUUCGGCGUAGUUUUCAGUAAAUACGA (((.((.....................((((....))))(((((((.......((((((((((((((......)))))))))))))).))))))))).)))................... ( -32.20) >DroSec_CAF1 27843 119 - 1 CGCGAAAU-UUUAGGAAAAAAUAAAUAAAGUUAGUGCUUAAACACCCAAAUAACGAUAUAUAUUCGCUCGACUGCGAAUAUGUAUCGCGGUGUUUUCGGCGUAGUUUUCAGUAAAUACGA .((..(((-((((............))))))).))(((.(((((((.......((((((((((((((......)))))))))))))).)))))))..)))(((.((......)).))).. ( -30.80) >DroSim_CAF1 31040 119 - 1 CGCGAAAU-UGUAGGAAAAAAUAAAUAAAGUUAGUGAUUAAACACCCAAAGAACGAUAUAUAUUCGCUCGACUGCGAAUAUGUAUCGCGGUGUUUUCGGCGUAGUUUUCAGUAAAUACGU ..((((.(-(((........))))................((((((.......((((((((((((((......)))))))))))))).))))))))))(((((.((......)).))))) ( -28.90) >DroEre_CAF1 25073 119 - 1 CGCGAAAUUGUUAGAAAAAAAUAAAUAGACUAAAUGCUUAAACAUCCAAAUAACGAUAUAUAUUCGCACGACUACGAAUAUGUAUCGCGGUGUU-UUGGCGUAGUUUUCAGUAAAUACGA ...(((.(((((.......)))))...(((((...(((.(((((((.......(((((((((((((........))))))))))))).))))))-).))).))))))))........... ( -26.90) >DroYak_CAF1 26802 120 - 1 CGCAAAAUUUUAAGGAAAAAAUAAAUAGAGUAAGUGCUUAAACAUCCAAAUAACGAUAUAUAUUCGCUCGACUUCGAAUAUGUAUCGCGGUGUUUUUGGCGUAGUUUUCAGCAAAUACGA .............................(((..((((.(((((((((((...(((((((((((((........))))))))))))).......))))).)).))))..))))..))).. ( -26.10) >consensus CGCGAAAUUUUUAGGAAAAAAUAAAUAAAGUUAGUGCUUAAACACCCAAAUAACGAUAUAUAUUCGCUCGACUGCGAAUAUGUAUCGCGGUGUUUUCGGCGUAGUUUUCAGUAAAUACGA (((........................((((....))))(((((((.......((((((((((((((......)))))))))))))).)))))))...)))................... (-23.84 = -24.36 + 0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:56 2006