| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,357,561 – 8,357,657 |
| Length | 96 |
| Max. P | 0.936142 |

| Location | 8,357,561 – 8,357,657 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 79.48 |
| Mean single sequence MFE | -20.93 |
| Consensus MFE | -15.43 |
| Energy contribution | -14.73 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8357561 96 + 22224390 UGCG------AA-AUAAUCCGA------UUGAAAACCCAGAUCCCGACAAUGAGAAAUUGUCGGGCCUACCACAUUCGACAACCGGUUCACUAAGCUCAAUCAUCACGA ....------..-.......((------((((..........(((((((((.....)))))))))..........(((.....)))..........))))))....... ( -20.80) >DroPse_CAF1 20084 99 + 1 -A----AAC--C-AUCCCCCGACCAUGCUCGU--AACCAGAUCCCGACAAUGAGAAGUUGUCGGGCCUACCACAUUCGACAACUGGUUCACUAAGCUCAAUCAUCACGA -.----...--.-.......((....(((.((--((((((..(((((((((.....))))))))).......(....)....)))))).))..)))....))....... ( -22.80) >DroGri_CAF1 2960 97 + 1 UACCC----AAAAAUU-CCCGC------UUG-UUGCACAGACCCCGACAAUGAGAAGUUGUCCGGCCUUCCCCAUUCAACAACUGGUUCACUAAGCUCAAUCAUCACGA .....----.......-...((------(((-.((..(((.....((((((.....)))))).((......)).........)))...)).)))))............. ( -15.20) >DroEre_CAF1 19942 94 + 1 --------CAAA-ACAAUCCGA------UCGAAAACCCAGAUCCCGACAAUGAGAAAUUGUCGGGCCUACCACAUUCGACAACCGGUUCACUAAGCUCAAUCAUCACGA --------....-.....(((.------(((((.........(((((((((.....))))))))).........)))))....)))....................... ( -20.17) >DroYak_CAF1 21384 100 + 1 CGAAUAAACAAA-ACA--CCGA------UCGAAAAAACAGAUCCCGACAAUGAGAAAUUGUCGGGCCUGCCACAUUCGACAACCGGUUCACUAAGCUCAAUCAUCACGA ............-..(--(((.------(((((....(((..(((((((((.....))))))))).))).....)))))....))))...................... ( -23.80) >DroPer_CAF1 20213 99 + 1 -A----AAC--C-AUACCCCGACCAUGCUCGU--AACCAGAUCCCGACAAUGAGAAGUUGUCGGGCCUACCACAUUCGACAACUGGUUCACUAAGCUCAAUCAUCACGA -.----...--.-.......((....(((.((--((((((..(((((((((.....))))))))).......(....)....)))))).))..)))....))....... ( -22.80) >consensus _A______C_AA_AUA_CCCGA______UCGAAAAACCAGAUCCCGACAAUGAGAAAUUGUCGGGCCUACCACAUUCGACAACCGGUUCACUAAGCUCAAUCAUCACGA ..................(((.......(((...........(((((((((.....)))))))))...........)))....)))....................... (-15.43 = -14.73 + -0.69)

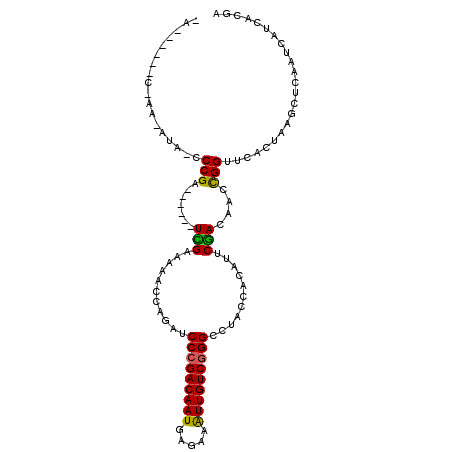
| Location | 8,357,561 – 8,357,657 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.48 |
| Mean single sequence MFE | -29.53 |
| Consensus MFE | -19.23 |
| Energy contribution | -19.07 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8357561 96 - 22224390 UCGUGAUGAUUGAGCUUAGUGAACCGGUUGUCGAAUGUGGUAGGCCCGACAAUUUCUCAUUGUCGGGAUCUGGGUUUUCAA------UCGGAUUAU-UU------CGCA .............((..(((((.(((((((..((((.(((....(((((((((.....)))))))))..))).)))).)))------)))).))))-).------.)). ( -33.20) >DroPse_CAF1 20084 99 - 1 UCGUGAUGAUUGAGCUUAGUGAACCAGUUGUCGAAUGUGGUAGGCCCGACAACUUCUCAUUGUCGGGAUCUGGUU--ACGAGCAUGGUCGGGGGAU-G--GUU----U- ((.((((.((...((((.((((.((((((..((....))..)).((((((((.......))))))))..))))))--)))))))).)))).))...-.--...----.- ( -28.20) >DroGri_CAF1 2960 97 - 1 UCGUGAUGAUUGAGCUUAGUGAACCAGUUGUUGAAUGGGGAAGGCCGGACAACUUCUCAUUGUCGGGGUCUGUGCAA-CAA------GCGGG-AAUUUUU----GGGUA .............((((((....((..((((((...(.(((...((.(((((.......))))).)).))).).)))-)))------..)).-.....))----)))). ( -21.40) >DroEre_CAF1 19942 94 - 1 UCGUGAUGAUUGAGCUUAGUGAACCGGUUGUCGAAUGUGGUAGGCCCGACAAUUUCUCAUUGUCGGGAUCUGGGUUUUCGA------UCGGAUUGU-UUUG-------- ..............(..((..(.(((((((..((((.(((....(((((((((.....)))))))))..))).)))).)))------)))).)..)-)..)-------- ( -27.50) >DroYak_CAF1 21384 100 - 1 UCGUGAUGAUUGAGCUUAGUGAACCGGUUGUCGAAUGUGGCAGGCCCGACAAUUUCUCAUUGUCGGGAUCUGUUUUUUCGA------UCGG--UGU-UUUGUUUAUUCG .......((.(((((.......((((...((((((...(((((((((((((((.....))))))))).))))))..)))))------))))--)..-...))))).)). ( -36.70) >DroPer_CAF1 20213 99 - 1 UCGUGAUGAUUGAGCUUAGUGAACCAGUUGUCGAAUGUGGUAGGCCCGACAACUUCUCAUUGUCGGGAUCUGGUU--ACGAGCAUGGUCGGGGUAU-G--GUU----U- (((((.((((((.((((.((((.((((((..((....))..)).((((((((.......))))))))..))))))--)))))).))))))...)))-)--)..----.- ( -30.20) >consensus UCGUGAUGAUUGAGCUUAGUGAACCAGUUGUCGAAUGUGGUAGGCCCGACAACUUCUCAUUGUCGGGAUCUGGGUUUUCGA______UCGGG_UAU_UU_G______U_ ((((((..((((..(.....)...))))..))).......((((((((((((.......)))))))).))))................))).................. (-19.23 = -19.07 + -0.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:51 2006