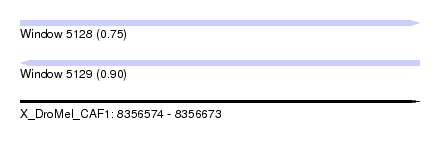
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,356,574 – 8,356,673 |
| Length | 99 |
| Max. P | 0.896522 |

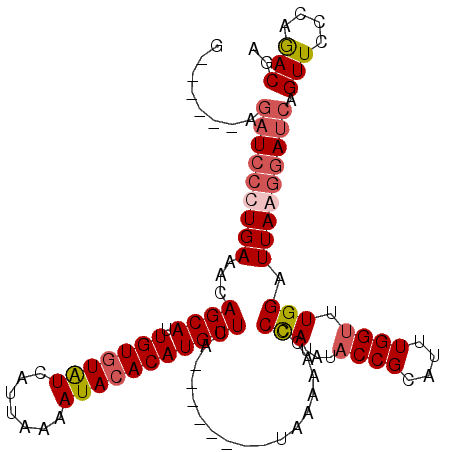
| Location | 8,356,574 – 8,356,673 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 83.81 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -19.12 |
| Energy contribution | -20.00 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8356574 99 + 22224390 UCGUCUGGGAACUGAUCCUUAAUCCAAACCAAAUGCGGUAUAUGGAAUUUUUA--------UAAGCAUGUGUAUUUUAAUGAUACACACUGCUGUUUCAAGGAUCU------C .............(((((((..((((.(((......)))...)))).......--------..((((((((((((.....)))))))).)))).....))))))).------. ( -28.10) >DroSec_CAF1 21896 99 + 1 UCGUUUGGAAACUGAUCCUUAAUCCAAACCAAAUGCGGUAUAUAGGGUUUUUA--------UAAGCAUGUGUAUUUCAAUGAUACACAAUGCUGUUUCAGGGAUCU------C ......(....).((((((((((((..(((......))).....)))))....--------..((((((((((((.....))))))).))))).....))))))).------. ( -27.00) >DroSim_CAF1 25091 99 + 1 UCGUCUGGAAACUGAUCCUUAAUCCAAACCAAAUGCGGUAUAUGGGGUUUUUA--------UAAGCAUGUGUAUUUUAAUGACACACAAUGCUGUUUCAGGGAUCU------C ......(....).((((((((((((..(((......))).....)))))....--------..(((((((((.((.....)).)))).))))).....))))))).------. ( -24.00) >DroEre_CAF1 18958 113 + 1 UCGUCUGAGAACUGAUCCUUAAUCCAAGCCAAAUGCGGUAUAUAGAGUUCUUAAGGUUUGCUUAGCAUGUGUAUUUUUAUGAUAGACAAUGCUGUUUCAAUUAGUUAAAGUGC .....(((((((((((.....)))...(((......)))......))))))))....(((..((((((((.((((.....)))).)).))))))...)))............. ( -20.20) >consensus UCGUCUGGAAACUGAUCCUUAAUCCAAACCAAAUGCGGUAUAUAGAGUUUUUA________UAAGCAUGUGUAUUUUAAUGAUACACAAUGCUGUUUCAAGGAUCU______C .............(((((((..((((.(((......)))...)))).................((((((((((((.....)))))))).)))).....)))))))........ (-19.12 = -20.00 + 0.88)

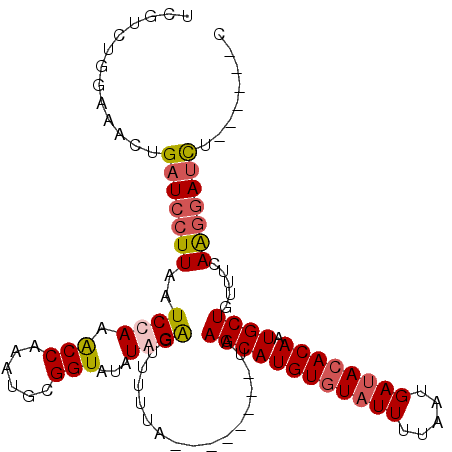
| Location | 8,356,574 – 8,356,673 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 83.81 |
| Mean single sequence MFE | -22.68 |
| Consensus MFE | -19.05 |
| Energy contribution | -20.43 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8356574 99 - 22224390 G------AGAUCCUUGAAACAGCAGUGUGUAUCAUUAAAAUACACAUGCUUA--------UAAAAAUUCCAUAUACCGCAUUUGGUUUGGAUUAAGGAUCAGUUCCCAGACGA .------.(((((((((...((((.(((((((.......)))))))))))..--------.......((((...((((....)))).)))))))))))))............. ( -29.40) >DroSec_CAF1 21896 99 - 1 G------AGAUCCCUGAAACAGCAUUGUGUAUCAUUGAAAUACACAUGCUUA--------UAAAAACCCUAUAUACCGCAUUUGGUUUGGAUUAAGGAUCAGUUUCCAAACGA .------.(((((.(((...(((((.((((((.......)))))))))))..--------........(((...((((....)))).))).))).)))))............. ( -21.70) >DroSim_CAF1 25091 99 - 1 G------AGAUCCCUGAAACAGCAUUGUGUGUCAUUAAAAUACACAUGCUUA--------UAAAAACCCCAUAUACCGCAUUUGGUUUGGAUUAAGGAUCAGUUUCCAGACGA .------.(((((.(((...(((((.((((((.......)))))))))))..--------........(((...((((....)))).))).))).))))).(((....))).. ( -24.40) >DroEre_CAF1 18958 113 - 1 GCACUUUAACUAAUUGAAACAGCAUUGUCUAUCAUAAAAAUACACAUGCUAAGCAAACCUUAAGAACUCUAUAUACCGCAUUUGGCUUGGAUUAAGGAUCAGUUCUCAGACGA ...........((((((...(((((.((.(((.......))).))))))).......((((((....((((....(((....)))..)))))))))).))))))......... ( -15.20) >consensus G______AGAUCCCUGAAACAGCAUUGUGUAUCAUUAAAAUACACAUGCUUA________UAAAAACCCCAUAUACCGCAUUUGGUUUGGAUUAAGGAUCAGUUCCCAGACGA ........(((((((((...((((.(((((((.......)))))))))))..................(((...((((....)))).))).))))))))).(((....))).. (-19.05 = -20.43 + 1.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:49 2006