
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,350,718 – 8,350,831 |
| Length | 113 |
| Max. P | 0.624270 |

| Location | 8,350,718 – 8,350,831 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.82 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -6.30 |
| Energy contribution | -7.48 |
| Covariance contribution | 1.18 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.24 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
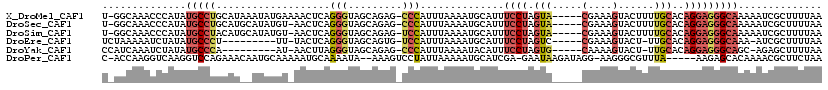
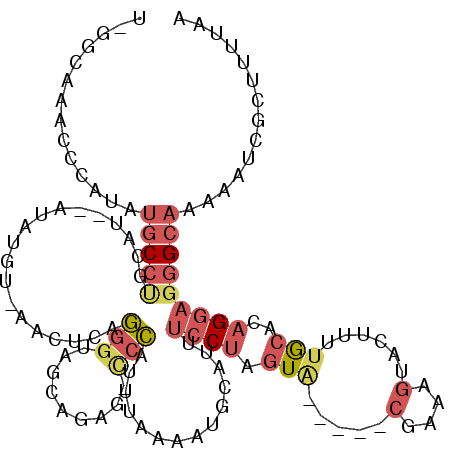
>X_DroMel_CAF1 8350718 113 + 22224390 U-GGCAAACCCAUAUGCCUGCAUAAAUAUGAAAACUCAGGGUAGCAGAG-CCCAUUUAAAAUGCAUUUCCUAGUA-----CGAAAGUACUUUUGCACAGGAGGGCAAAAAUCGCUUUUAA .-(((..((((.((((....))))....(((....)))))))......(-(((.(((....((((......((((-----(....)))))..))))..))))))).......)))..... ( -29.50) >DroSec_CAF1 16101 112 + 1 U-GGCAAACCCAUAUGCCUGCAUGCAUAUGU-AACUCAGGGUAGCAGAG-CCCAUUUAAAAUGCAUUUCCUAGUA-----CGAAAGUACUUUUGCACAGGAGGGCAAAAAUCGCUUUUAA .-(((.....(((((((......))))))).-......((((......)-)))........(((.((((((((((-----(....))))).......)))))))))......)))..... ( -30.91) >DroSim_CAF1 19117 112 + 1 U-GGCAAACCCAUAUGCCUACAUGCAUAUGU-AACUCAGGGUAGCAGAG-UCCAUUUAAAAUGCAUUUCCUAGUA-----CGAAAGUACUUUUGCACAGGAGGGCAAAAAUCGCUUUUAA (-((((((..(((((((......))))))).-.....((((..(((...-...........)))...))))((((-----(....)))))))))).)).((((((.......)))))).. ( -27.94) >DroEre_CAF1 13281 102 + 1 UCUAAAAAUCUAUAUGCCCU---------UU-UACUCAGGGUAGCAGUG-UCCAUUUAAAAUGCAUUUCCUAGUC-----CGAAAGUACU-UUGCACAGGAGGGCAAA-AUCGCUUUUAA ..............((((((---------((-(...((((((((((.((-......))...)))...........-----(....)))))-)))...)))))))))..-........... ( -21.90) >DroYak_CAF1 14002 102 + 1 CCAUCAAAUCUAUAUGCCCA---------AU-AACUUAGGGUAGCAGAG-CCCAUUUAAAAUACAUUUCCUAGUG-----CAAAAGUACU-UUGCACAGGAGGGCAGC-AGAGCUUUUAA ........(((...(((((.---------..-......((((......)-)))..............((((.(((-----((((.....)-)))))))))))))))..-)))........ ( -31.10) >DroPer_CAF1 12527 110 + 1 C-ACCAAGGUCAAGGUCCAGAAACAAUGCAAAAAUGCAAAAUA--AAAGUCCUAUUAAAAAUGCAUCGA-GAAUAAGAUAGG-AAGGGCGUUUA-----AAGAGCACAAAACGCUUCUAA .-.........((.((((........((((....)))).....--....(((((((.............-......))))))-).)))).))..-----..((((.......)))).... ( -16.91) >consensus U_GGCAAACCCAUAUGCCUGCAU__AUAUGU_AACUCAGGGUAGCAGAG_CCCAUUUAAAAUGCAUUUCCUAGUA_____CGAAAGUACUUUUGCACAGGAGGGCAAAAAUCGCUUUUAA ..............(((((...................(((.........)))..............((((.(((.....(....)......)))..))))))))).............. ( -6.30 = -7.48 + 1.18)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:45 2006