
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,348,584 – 8,348,674 |
| Length | 90 |
| Max. P | 0.660140 |

| Location | 8,348,584 – 8,348,674 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 62.15 |
| Mean single sequence MFE | -16.67 |
| Consensus MFE | -5.36 |
| Energy contribution | -5.87 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8348584 90 + 22224390 AAGAGCAACCUUAACUGACCAAAA----CAAAAAAAAUGUGUAUGAAUAUAUAAAUAAAGCUUUAC------------AAACGUCGAAAGGCUGAAAAGCUUUGGG ..................((....----.........((((((....))))))...((((((((.(------------(...(((....))))).)))))))))). ( -16.10) >DroPse_CAF1 10517 96 + 1 UAAUGCAACCUUAACGGACGGACAGUGA--AAAAAAAUGCGAAACAAAUCGAACGGCA--GCGAACCGAUGUGAAUUGGGAGCCCAGA------AAACGCUCUUAA ...((((.((.....))((.....))..--.......))))...((.((((..((...--.))...)))).))..((((((((.....------....)))))))) ( -14.10) >DroSec_CAF1 14013 86 + 1 AAGAGCAACCUUAACUGACCAAAA------A--AAAAUGUGUAUGAAUAUAUAAAUAAAGCUUUAC------------AAGCGUCGAAAGACUGAAAAGCUUUGGG ..................((....------.--....((((((....))))))...((((((((.(------------(...(((....))))).)))))))))). ( -16.70) >DroSim_CAF1 16987 88 + 1 AAGAGCAACCUUAACUGACCAAAA------AAAAAAAUGUGUAUGAAUAUAUAAAUAAAGCUUUAC------------AAGCGUCGAAAGGCUGAAAAGCUUUGGG ..................((....------.......((((((....))))))...((((((((.(------------(...(((....))))).)))))))))). ( -16.10) >DroEre_CAF1 11133 83 + 1 AGGAGUAACCUUAACUGACCAAA----------AAAAUGUGUAUGAAUA-AUAAAUAAAGCUUUAC------------ACAAGUCGAAAGACUGCAGAGCUUUGGG (((.....))).......(((((----------....((((((......-.............)))------------)))((((....)))).......))))). ( -16.51) >DroAna_CAF1 976 95 + 1 AAGA---AUCUGAAAUCAGAAAUC--------AGAAAUGCGAAUCAAAUUGAACCGAAAACCGUGCCGCAGUGAAUCGAAAGGCUGAAAAGCUGAAAAGCUUUUAA ....---.((((....))))..((--------((.....(((.(((...((..(((.....)).)...)).))).))).....))))((((((....))))))... ( -20.50) >consensus AAGAGCAACCUUAACUGACCAAAA______A_AAAAAUGUGUAUGAAUAUAUAAAUAAAGCUUUAC____________AAACGUCGAAAGGCUGAAAAGCUUUGGG .(((((...............................((((((....)))))).............................(((....)))......)))))... ( -5.36 = -5.87 + 0.51)

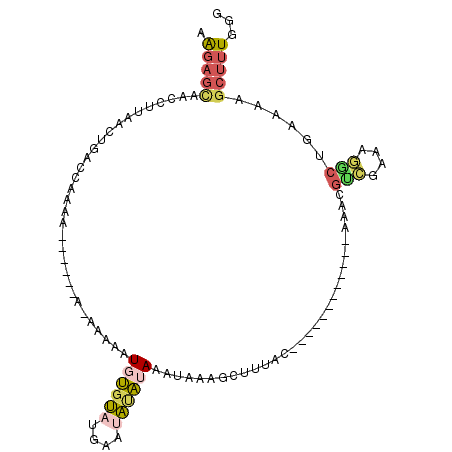
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:42 2006