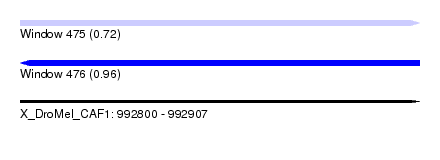
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 992,800 – 992,907 |
| Length | 107 |
| Max. P | 0.959794 |

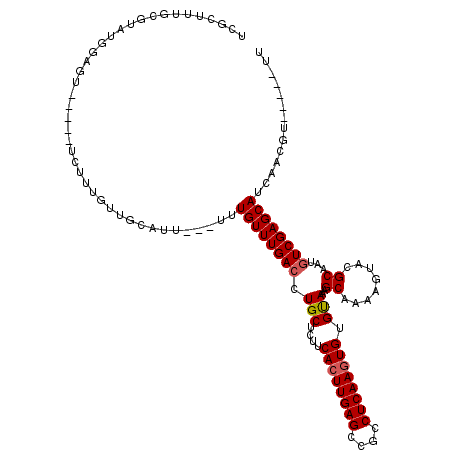
| Location | 992,800 – 992,907 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.25 |
| Mean single sequence MFE | -30.26 |
| Consensus MFE | -20.78 |
| Energy contribution | -21.02 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 992800 107 + 22224390 UCGUUUUGCGUAUGGAGU-----UCUUUGUUGCGUU---UUUGUUUGACCUGCUCUUCACUUGAGCAGCCUCAAGUGUGUAAAGCAAAAGUGCGCAAUGUCGAGCAUCAACGU-----UU .......((((.((..((-----((..(((((((((---(((((((....(((....((((((((....)))))))).)))))))))))..))))))))..))))..))))))-----.. ( -34.50) >DroSec_CAF1 75332 104 + 1 UCGCUUUGCGUAUGGAGU-----UCUUUGUUGCCUU---UUUGUUUGACUUGCUCUUCACUUGAGCCGCCUCAAGUGUGUAAAGCAAAAGUUAGCAAUGUCGAGCAUCAACG-------- ........(((.((..((-----((..((((((.((---(((((((....(((....((((((((....)))))))).))))))))))))...))))))..))))..)))))-------- ( -28.50) >DroSim_CAF1 54700 107 + 1 UCGCUUUGCGUAUGGAGU-----UCUUUGUUGCGUU---UUUGUUUGACCUGCUCUUCACUUGAGCCGCCUCAAGUGUGUAAAGCAAAAGUGCGCAAUGUCGAGCAUCAACGU-----UU .......((((.((..((-----((..(((((((((---(((((((....(((....((((((((....)))))))).)))))))))))..))))))))..))))..))))))-----.. ( -34.50) >DroEre_CAF1 88820 96 + 1 UUGCUUUAUGUC---------------------AUU---UUUGUUUGACCUGCUCUUCACUUGAGGCGCCUCAAGUGUGUAAAGCAAAAGUACGCAAUGUCGAGCAUCAACAUCAACGUU ((((((((((((---------------------(..---......))))........((((((((....)))))))).)))))))))....(((..((((.(.....).))))...))). ( -26.20) >DroYak_CAF1 81245 111 + 1 UCGCUUUGCGUUUGGAGUUCAGUUCUUUGUUGUAUUUGUGUUGUUUGACCUGCUCUUCAGUUGAGGCGCCUCAAGUGUGCAAAGCAAAAGUACGCAAUCUCGAGCAUCAGC--------- ..(((.(((....((((.......)))).(((...((((((..((((...(((....((.(((((....))))).)).)))...))))...))))))...))))))..)))--------- ( -27.60) >consensus UCGCUUUGCGUAUGGAGU_____UCUUUGUUGCAUU___UUUGUUUGACCUGCUCUUCACUUGAGCCGCCUCAAGUGUGUAAAGCAAAAGUACGCAAUGUCGAGCAUCAACGU_____UU .........................................((((((((.(((....((((((((....)))))))).)))..((........))...)))))))).............. (-20.78 = -21.02 + 0.24)

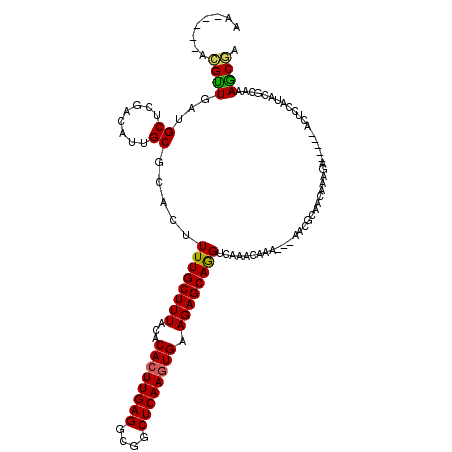
| Location | 992,800 – 992,907 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.25 |
| Mean single sequence MFE | -27.26 |
| Consensus MFE | -21.18 |
| Energy contribution | -20.86 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 992800 107 - 22224390 AA-----ACGUUGAUGCUCGACAUUGCGCACUUUUGCUUUACACACUUGAGGCUGCUCAAGUGAAGAGCAGGUCAAACAAA---AACGCAACAAAGA-----ACUCCAUACGCAAAACGA ..-----.((((..(((..(((.(((((((....)))......((((((((....))))))))....))))))).......---..(........).-----.........))).)))). ( -26.40) >DroSec_CAF1 75332 104 - 1 --------CGUUGAUGCUCGACAUUGCUAACUUUUGCUUUACACACUUGAGGCGGCUCAAGUGAAGAGCAAGUCAAACAAA---AAGGCAACAAAGA-----ACUCCAUACGCAAAGCGA --------((((..(((..(((.(((((......((.....))((((((((....))))))))...)))))))).......---..(....).....-----.........))).)))). ( -26.20) >DroSim_CAF1 54700 107 - 1 AA-----ACGUUGAUGCUCGACAUUGCGCACUUUUGCUUUACACACUUGAGGCGGCUCAAGUGAAGAGCAGGUCAAACAAA---AACGCAACAAAGA-----ACUCCAUACGCAAAGCGA ..-----.((((..(((..(((.(((((((....)))......((((((((....))))))))....))))))).......---..(........).-----.........))).)))). ( -26.40) >DroEre_CAF1 88820 96 - 1 AACGUUGAUGUUGAUGCUCGACAUUGCGUACUUUUGCUUUACACACUUGAGGCGCCUCAAGUGAAGAGCAGGUCAAACAAA---AAU---------------------GACAUAAAGCAA .((((.(((((((.....)))))))))))....((((((((..((((((((....))))))))........((((......---..)---------------------))).)))))))) ( -32.30) >DroYak_CAF1 81245 111 - 1 ---------GCUGAUGCUCGAGAUUGCGUACUUUUGCUUUGCACACUUGAGGCGCCUCAACUGAAGAGCAGGUCAAACAACACAAAUACAACAAAGAACUGAACUCCAAACGCAAAGCGA ---------....((((........))))....(((((((((.((.(((((....))))).))..((((((.((.....................)).)))..))).....))))))))) ( -25.00) >consensus AA_____ACGUUGAUGCUCGACAUUGCGCACUUUUGCUUUACACACUUGAGGCGGCUCAAGUGAAGAGCAGGUCAAACAAA___AACGCAACAAAGA_____ACUCCAUACGCAAAGCGA ........((((...((........)).....((((((((...((((((((....)))))))).))))))))...........................................)))). (-21.18 = -20.86 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:29 2006