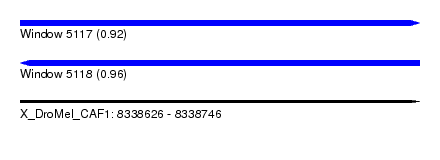
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,338,626 – 8,338,746 |
| Length | 120 |
| Max. P | 0.960302 |

| Location | 8,338,626 – 8,338,746 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.58 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -23.44 |
| Energy contribution | -23.20 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
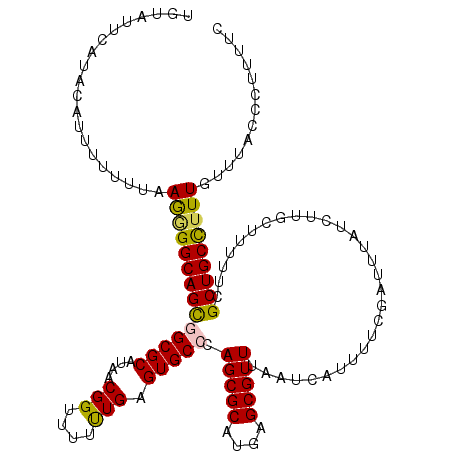
>X_DroMel_CAF1 8338626 120 + 22224390 UGUAUUCAUAUAAUUUUUUAAAAGGCAGCGGCGCAUAACGGUUUUUUGAGUGCCCAGCGCAUGAGCGUUUAAUCAUUUUCGAUUUAUCUUGCUUUUUCGCUGCCUUUGUUUACCCUUUUU ....................((((((((((((((.....(((.........)))..))))..(((((...((((......)))).....)))))...))))))))))............. ( -30.50) >DroSec_CAF1 3482 120 + 1 UUUAUUCAUACGUUUUUUUAAGGGGCAGUAGCGCAUAACGGUUUUCUGAGUGCCCAGCGCAUGAGCGUUUAAUCAUUUUCGAUUUAUCUGGCUUUUUCGCUGCCUUUGUUUACCCUUUUC ...................(((((((((..((((((..(((....))).))))..((((...((((....((((......))))......))))...))))))..))))...)))))... ( -25.00) >DroSim_CAF1 3568 120 + 1 UUUAUUCAUACAUUUUUUUAAGGGGCAGCAGCGCAUAACGGUUUUCUGAGUGCCCAGCGCAUGAGCGUUUAAUCAUUUUCGAUUUAUCUGGCUUUUUCGCUGCCUUUGUUUACCCUUUUC ....................(((((((((.((((....(((....))).))))(((((((....)))...((((......))))...)))).......)))))))))............. ( -26.70) >DroEre_CAF1 1627 114 + 1 ------CAUAUAUUUUUUUAAGGGGCAGCGGCGCAUAACGGGUUUUUGAGUGCCCAGCGCAUGAGCGUUUAAUCAUUUUCAAUUUAUCUUGGUUUUUCGCUGCCUUUGUUUACCCUUUUU ------..............((((((((((((((.....((((........)))).))))((((........)))).....................))))))))))............. ( -29.40) >DroYak_CAF1 2226 119 + 1 UGUUUUAAUACAUU-UUUUAAGGGGCAGCGGCGCAUAACGGGUUUUUGAGUGCCCAGCGCAUGAGCGUUUAAUCAUUUUCGAUUUAUCUUGGUUUUUCGCUGCUAUUGUUUACCCUUUUC ..............-....((((((((((((((...(((((((........))))(((((....)))))......................)))...))))))...)))...)))))... ( -28.50) >consensus UGUAUUCAUACAUUUUUUUAAGGGGCAGCGGCGCAUAACGGUUUUUUGAGUGCCCAGCGCAUGAGCGUUUAAUCAUUUUCGAUUUAUCUUGCUUUUUCGCUGCCUUUGUUUACCCUUUUC ....................((((((((((((((....(((....))).))))).(((((....))))).............................)))))))))............. (-23.44 = -23.20 + -0.24)


| Location | 8,338,626 – 8,338,746 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.58 |
| Mean single sequence MFE | -24.76 |
| Consensus MFE | -20.46 |
| Energy contribution | -20.10 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
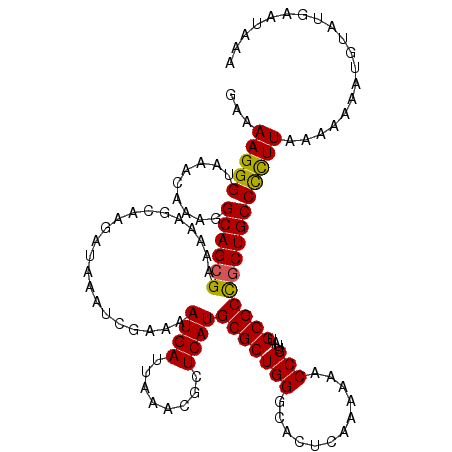
>X_DroMel_CAF1 8338626 120 - 22224390 AAAAAGGGUAAACAAAGGCAGCGAAAAAGCAAGAUAAAUCGAAAAUGAUUAAACGCUCAUGCGCUGGGCACUCAAAAAACCGUUAUGCGCCGCUGCCUUUUAAAAAAUUAUAUGAAUACA .............((((((((((.....(((.(((.((((......))))....(((((.....)))))............))).)))..)))))))))).................... ( -26.70) >DroSec_CAF1 3482 120 - 1 GAAAAGGGUAAACAAAGGCAGCGAAAAAGCCAGAUAAAUCGAAAAUGAUUAAACGCUCAUGCGCUGGGCACUCAGAAAACCGUUAUGCGCUACUGCCCCUUAAAAAAACGUAUGAAUAAA ...(((((....)...(((((........((((...((((......))))...(((....)))))))(((....(.....)....)))....)))))))))................... ( -20.60) >DroSim_CAF1 3568 120 - 1 GAAAAGGGUAAACAAAGGCAGCGAAAAAGCCAGAUAAAUCGAAAAUGAUUAAACGCUCAUGCGCUGGGCACUCAGAAAACCGUUAUGCGCUGCUGCCCCUUAAAAAAAUGUAUGAAUAAA ...(((((........((((((.......((((...((((......))))...(((....)))))))(((....(.....)....)))))))))..)))))................... ( -26.10) >DroEre_CAF1 1627 114 - 1 AAAAAGGGUAAACAAAGGCAGCGAAAAACCAAGAUAAAUUGAAAAUGAUUAAACGCUCAUGCGCUGGGCACUCAAAAACCCGUUAUGCGCCGCUGCCCCUUAAAAAAAUAUAUG------ ...(((((....)...(((((((.....................((((........))))((((((((..........))))....))))))))))))))).............------ ( -25.60) >DroYak_CAF1 2226 119 - 1 GAAAAGGGUAAACAAUAGCAGCGAAAAACCAAGAUAAAUCGAAAAUGAUUAAACGCUCAUGCGCUGGGCACUCAAAAACCCGUUAUGCGCCGCUGCCCCUUAAAA-AAUGUAUUAAAACA ...(((((.........((((((.....................((((........))))((((((((..........))))....)))))))))))))))....-.............. ( -24.80) >consensus GAAAAGGGUAAACAAAGGCAGCGAAAAAGCAAGAUAAAUCGAAAAUGAUUAAACGCUCAUGCGCUGGGCACUCAAAAAACCGUUAUGCGCCGCUGCCCCUUAAAAAAAUGUAUGAAUAAA ...(((((.........((((((.....................((((........))))(((((((............)))....)))))))))))))))................... (-20.46 = -20.10 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:39 2006