
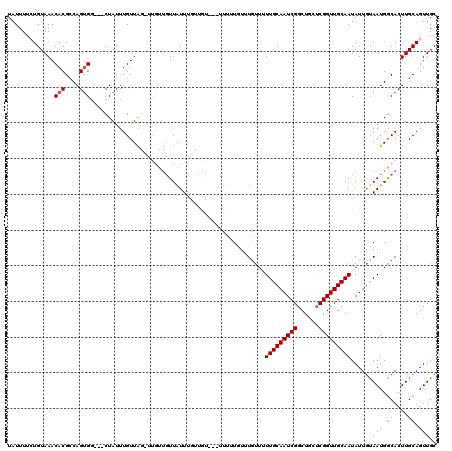
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,338,016 – 8,338,173 |
| Length | 157 |
| Max. P | 0.999428 |

| Location | 8,338,016 – 8,338,133 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.52 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -15.88 |
| Energy contribution | -16.24 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8338016 117 + 22224390 UAUUUUCUGUAAACACGCCAGCGU---CUAUUUGUUAGCUUGUUGUUAUUUGUUGUGUGGUUUUAUUUUUUGUUGCAAUCGGCUGCUCGGUUGCAAUAUUAUAAUGACACUUGCAGUUGC ......(((((((((.((.((((.---.....)))).)).))))..........((((.(((.((.....((((((((((((....)))))))))))).)).))).)))).))))).... ( -30.80) >DroSec_CAF1 2891 117 + 1 UAUUUUCUGUAAACACGCCAGUGG---CUAUUUGUUAGUUCGUUGUUAUUUGUUGUAAUUUUUUUUUUGUUUUUGCAAUCGGCUGCUCGGUUGCAAUAUUGUAAUGGCACUUGCAGUUGC ......((((((.(((....)))(---(((((...((((.....(((((.....))))).............((((((((((....)))))))))).)))).))))))..)))))).... ( -24.60) >DroSim_CAF1 2973 115 + 1 UAUUUUCUGUAAACACGCCAGUGG---CUAUUUGUUAGUUUGUUGUUAUUUGUUGUA--UUUUUUUUUGUUUUUGCAAUCGGCUUCUCGGUUGCAAUAUUGUAAUGGCACUUGCAGUUGC ......((((((.(((....)))(---(((.....)))).(((..((((........--.............((((((((((....))))))))))....))))..))).)))))).... ( -24.01) >DroEre_CAF1 984 106 + 1 UAUUUUGUGUAAACACGACAGUGG---CUAUUUGUUA--UUAUUGUUGUU---------UUUGUGUGUGUUUUUGCAAUCGGCUGUUCGGUUGCAAUAUUGCAAUGGCACUUGCAGUUGC ......(..((((.((((((((((---.((.....))--)))))))))).---------))))..)......((((((((((....))))))))))....(((((.((....)).))))) ( -28.10) >DroYak_CAF1 1617 103 + 1 UAUUUUGUGUAAACACGCCAGUGGCGGCUAUUUGUUG--------UUAUU---------UUUGUGUGUGCUUUUGCAAUCGGCUGUGCGGUUGCAAUAUUGCAAUGGCACUUGCAGUUGC ........(((.(((((..(((((((((.....))))--------)))))---------..))))).)))....(((((..((.((((.((((((....)))))).))))..)).))))) ( -36.10) >consensus UAUUUUCUGUAAACACGCCAGUGG___CUAUUUGUUAG_UUGUUGUUAUUUGUUGU___UUUUUGUUUGUUUUUGCAAUCGGCUGCUCGGUUGCAAUAUUGUAAUGGCACUUGCAGUUGC .............(((....))).................................................(((((((((......)))))))))....(((((.((....)).))))) (-15.88 = -16.24 + 0.36)


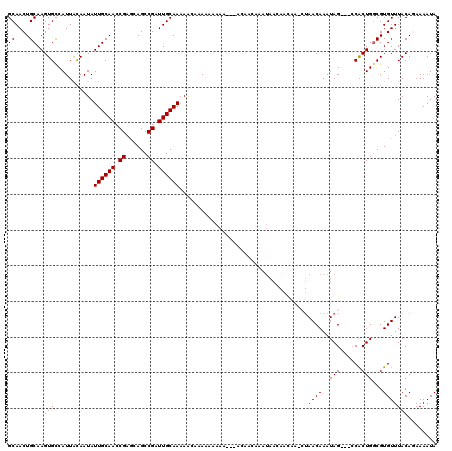
| Location | 8,338,016 – 8,338,133 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.52 |
| Mean single sequence MFE | -18.23 |
| Consensus MFE | -10.42 |
| Energy contribution | -10.42 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8338016 117 - 22224390 GCAACUGCAAGUGUCAUUAUAAUAUUGCAACCGAGCAGCCGAUUGCAACAAAAAAUAAAACCACACAACAAAUAACAACAAGCUAACAAAUAG---ACGCUGGCGUGUUUACAGAAAAUA ..((((((.((((((.........((((((.((......)).)))))).................................(....).....)---))))).))).)))........... ( -17.40) >DroSec_CAF1 2891 117 - 1 GCAACUGCAAGUGCCAUUACAAUAUUGCAACCGAGCAGCCGAUUGCAAAAACAAAAAAAAAAUUACAACAAAUAACAACGAACUAACAAAUAG---CCACUGGCGUGUUUACAGAAAAUA ....(((.(((..(..........((((((.((......)).))))))............................................(---((...))))..))).)))...... ( -16.50) >DroSim_CAF1 2973 115 - 1 GCAACUGCAAGUGCCAUUACAAUAUUGCAACCGAGAAGCCGAUUGCAAAAACAAAAAAAAA--UACAACAAAUAACAACAAACUAACAAAUAG---CCACUGGCGUGUUUACAGAAAAUA ....(((.(((..(..........((((((.((......)).)))))).............--.............................(---((...))))..))).)))...... ( -16.50) >DroEre_CAF1 984 106 - 1 GCAACUGCAAGUGCCAUUGCAAUAUUGCAACCGAACAGCCGAUUGCAAAAACACACACAAA---------AACAACAAUAA--UAACAAAUAG---CCACUGUCGUGUUUACACAAAAUA ((((..((....))..))))....((((((.((......)).))))))(((((((((....---------...........--..........---....))).)))))).......... ( -17.25) >DroYak_CAF1 1617 103 - 1 GCAACUGCAAGUGCCAUUGCAAUAUUGCAACCGCACAGCCGAUUGCAAAAGCACACACAAA---------AAUAA--------CAACAAAUAGCCGCCACUGGCGUGUUUACACAAAAUA ((....))..((((..(((((((..(((....)))......)))))))..)))).......---------.....--------.........((((....))))(((....)))...... ( -23.50) >consensus GCAACUGCAAGUGCCAUUACAAUAUUGCAACCGAGCAGCCGAUUGCAAAAACAAAAAAAAA___ACAACAAAUAACAACAA_CUAACAAAUAG___CCACUGGCGUGUUUACAGAAAAUA ((....))..(((...........((((((.((......)).))))))....................................((((..(((......)))...)))))))........ (-10.42 = -10.42 + -0.00)

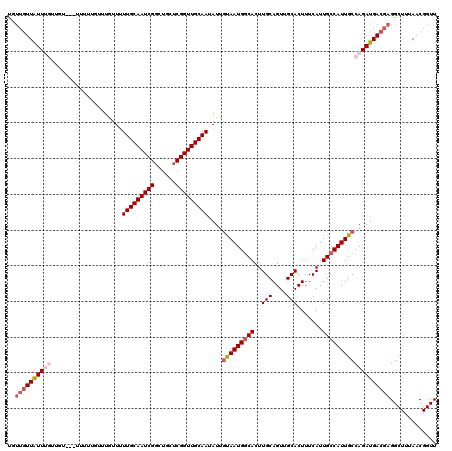
| Location | 8,338,053 – 8,338,173 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.19 |
| Mean single sequence MFE | -33.34 |
| Consensus MFE | -26.84 |
| Energy contribution | -28.24 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8338053 120 + 22224390 UGUUGUUAUUUGUUGUGUGGUUUUAUUUUUUGUUGCAAUCGGCUGCUCGGUUGCAAUAUUAUAAUGACACUUGCAGUUGCACUUUCAUUGCCAUUGCCAGAUGACGAGGCUUUAACGGUU ..((((((((((..((((.(((.((.....((((((((((((....)))))))))))).)).))).))))..(((((.(((.......))).)))))))))))))))............. ( -35.50) >DroSec_CAF1 2928 120 + 1 CGUUGUUAUUUGUUGUAAUUUUUUUUUUGUUUUUGCAAUCGGCUGCUCGGUUGCAAUAUUGUAAUGGCACUUGCAGUUGCACUUUCAUUGCCAUUGCCAGAUGACGAGGCUUUAACGGUU (.((((((((((....................((((((((((....))))))))))....(((((((((..(((....))).......))))))))))))))))))).)........... ( -33.70) >DroSim_CAF1 3010 118 + 1 UGUUGUUAUUUGUUGUA--UUUUUUUUUGUUUUUGCAAUCGGCUUCUCGGUUGCAAUAUUGUAAUGGCACUUGCAGUUGCACUUUCAUUGCCAUUGCCAGAUGACGAGGCUUUAACGGUU ..((((((((((.....--.............((((((((((....))))))))))....(((((((((..(((....))).......)))))))))))))))))))............. ( -32.80) >DroEre_CAF1 1019 111 + 1 UAUUGUUGUU---------UUUGUGUGUGUUUUUGCAAUCGGCUGUUCGGUUGCAAUAUUGCAAUGGCACUUGCAGUUGCACUUUCAUUGCCAUUGCCAGAUGACGAGGCUUUAACGGUU .(((((((.(---------(((((((.((...((((((((((....))))))))))....(((((((((..(((....))).......))))))))))).)).))))))...))))))). ( -32.90) >DroYak_CAF1 1654 106 + 1 -----UUAUU---------UUUGUGUGUGCUUUUGCAAUCGGCUGUGCGGUUGCAAUAUUGCAAUGGCACUUGCAGUUGCACUUUCAUUGCCAUUGCCAGAUGACGAGGCUUUAACGGUU -----.....---------...(((.((((..((((((......((((.((((((....)))))).))))))))))..))))...))).(((.(((.(....).)))))).......... ( -31.80) >consensus UGUUGUUAUUUGUUGU___UUUUUGUUUGUUUUUGCAAUCGGCUGCUCGGUUGCAAUAUUGUAAUGGCACUUGCAGUUGCACUUUCAUUGCCAUUGCCAGAUGACGAGGCUUUAACGGUU ..((((((((((....................(((((((((......)))))))))....(((((((((..(((....))).......)))))))))))))))))))............. (-26.84 = -28.24 + 1.40)


| Location | 8,338,053 – 8,338,173 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.19 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -22.18 |
| Energy contribution | -22.42 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.59 |
| SVM RNA-class probability | 0.999428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8338053 120 - 22224390 AACCGUUAAAGCCUCGUCAUCUGGCAAUGGCAAUGAAAGUGCAACUGCAAGUGUCAUUAUAAUAUUGCAACCGAGCAGCCGAUUGCAACAAAAAAUAAAACCACACAACAAAUAACAACA ....((((..(((..(((....)))...)))(((((...(((....)))....)))))......((((((.((......)).))))))........................)))).... ( -23.10) >DroSec_CAF1 2928 120 - 1 AACCGUUAAAGCCUCGUCAUCUGGCAAUGGCAAUGAAAGUGCAACUGCAAGUGCCAUUACAAUAUUGCAACCGAGCAGCCGAUUGCAAAAACAAAAAAAAAAUUACAACAAAUAACAACG ....((((..(((.........)))(((((((.......(((....)))..)))))))......((((((.((......)).))))))........................)))).... ( -24.60) >DroSim_CAF1 3010 118 - 1 AACCGUUAAAGCCUCGUCAUCUGGCAAUGGCAAUGAAAGUGCAACUGCAAGUGCCAUUACAAUAUUGCAACCGAGAAGCCGAUUGCAAAAACAAAAAAAAA--UACAACAAAUAACAACA ....((((..(((.........)))(((((((.......(((....)))..)))))))......((((((.((......)).)))))).............--.........)))).... ( -24.60) >DroEre_CAF1 1019 111 - 1 AACCGUUAAAGCCUCGUCAUCUGGCAAUGGCAAUGAAAGUGCAACUGCAAGUGCCAUUGCAAUAUUGCAACCGAACAGCCGAUUGCAAAAACACACACAAA---------AACAACAAUA ....(((................(((((((((.......(((....)))..)))))))))....((((((.((......)).)))))).))).........---------.......... ( -26.10) >DroYak_CAF1 1654 106 - 1 AACCGUUAAAGCCUCGUCAUCUGGCAAUGGCAAUGAAAGUGCAACUGCAAGUGCCAUUGCAAUAUUGCAACCGCACAGCCGAUUGCAAAAGCACACACAAA---------AAUAA----- ..........(((..(((....)))...))).......((((...(((((((((..(((((....)))))..))))......)))))...)))).......---------.....----- ( -26.60) >consensus AACCGUUAAAGCCUCGUCAUCUGGCAAUGGCAAUGAAAGUGCAACUGCAAGUGCCAUUACAAUAUUGCAACCGAGCAGCCGAUUGCAAAAACAAAAAAAAA___ACAACAAAUAACAACA ....((((..(((.........)))(((((((.......(((....)))..)))))))......((((((.((......)).))))))........................)))).... (-22.18 = -22.42 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:36 2006