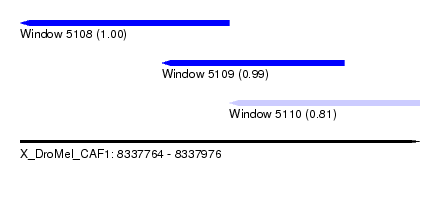
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,337,764 – 8,337,976 |
| Length | 212 |
| Max. P | 0.998184 |

| Location | 8,337,764 – 8,337,875 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.84 |
| Mean single sequence MFE | -21.63 |
| Consensus MFE | -19.04 |
| Energy contribution | -19.20 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.998184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8337764 111 - 22224390 --UUAGCAUAAUGCAAUUAGUGUAAAAAAUCAAA--CAAACUA---UAAAUUAAUACUUAGCAUUACAUUAAUUGCU--GCGAAAUUCCUCUUUACUGCGUUCUAAAAAUAGCGUGCAGC --...(((....(((((((((((((.........--.......---.................))))))))))))))--))..............(((((..(((....)))..))))). ( -21.51) >DroSec_CAF1 2638 110 - 1 --UUAGCAUAAUGCAAUUAGUGUAAA-AAUCAAA--CAAACUA---UAAAUUAAUACUUAGCAUUACAUUAAUUGCU--GCGAAAUUCCCCUUUACUGCGUUCUAAAAAUAGCGUGCAGC --...(((....(((((((((((((.-.......--.......---.................))))))))))))))--))..............(((((..(((....)))..))))). ( -21.55) >DroSim_CAF1 2721 110 - 1 --UUAGCAUAAUGCAAUUAGUGUAAA-AAUCAAA--CAAACUA---UAAAUUAAUACUUAGCAUUACAUUAAUUGCU--GCGAAAUUCCCCUUUACUGCGUUCUAAAAAUAGCGUGCAGC --...(((....(((((((((((((.-.......--.......---.................))))))))))))))--))..............(((((..(((....)))..))))). ( -21.55) >DroEre_CAF1 718 119 - 1 UGUUAGCAUAAUGCAAUUAGUGUAAA-AAUCUAAACUAAACUAUCUAAAAUUAAUACUUAGCAUUACAUUAAUUGCCCCGCGAUAUGCCUCUUUACUGCUCUCUAAAAAUAGCGUGCAGC .....(((((..(((((((((((((.-...((((.......................))))..))))))))))))).......))))).......((((.(.(((....))).).)))). ( -22.10) >DroYak_CAF1 1348 112 - 1 UGUUAGCAUAAUGCAAUUAGCGUAAA-AAUCAAA--CAAACAA---AAAAUUAAUACUUAGCAUUACAUUAAUUGCU--GCGGGAUGCCACUUUACUGCGCUCUAAAAAUAGCGUGCAGC .....((((...((((((((.((((.-.......--.......---.................)))).)))))))).--.....)))).......((((((.(((....))).)))))). ( -21.45) >consensus __UUAGCAUAAUGCAAUUAGUGUAAA_AAUCAAA__CAAACUA___UAAAUUAAUACUUAGCAUUACAUUAAUUGCU__GCGAAAUUCCCCUUUACUGCGUUCUAAAAAUAGCGUGCAGC .....((.....(((((((((((((......................................)))))))))))))...))..............((((((.(((....))).)))))). (-19.04 = -19.20 + 0.16)

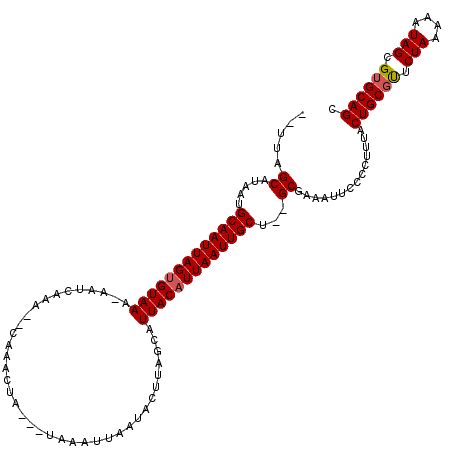
| Location | 8,337,839 – 8,337,936 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.60 |
| Mean single sequence MFE | -31.94 |
| Consensus MFE | -18.96 |
| Energy contribution | -19.36 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.14 |
| Mean z-score | -4.19 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8337839 97 - 22224390 A---CA----------CUGCUUGCACGCUGGCCUUAAACUUUGCGUU-UCAGGGCCACGUUA-GCAAAA-CCAAUUG-----UUAGCAUAAUGCAAUUAGUGUAAAAAAUCAAA--CAAA (---((----------(((.(((((.(.(((((((((((.....)))-).))))))))((((-((((..-....)))-----)))))....))))).))))))...........--.... ( -32.20) >DroSec_CAF1 2713 107 - 1 A---CACUGCACUGCACUGCUUGCACGCUGGCCUUAAACUUUGCGUU-UCAGGGCCACGCUA-GCAAAAAAAAAUUG-----UUAGCAUAAUGCAAUUAGUGUAAA-AAUCAAA--CAAA .---........(((((((.(((((.(.(((((((((((.....)))-).))))))))((((-((((.......)))-----)))))....))))).)))))))..-.......--.... ( -35.60) >DroSim_CAF1 2796 106 - 1 A---CACUGCACUGCACUGCUUGCACGCUGGCCUUAAACUUUGCGUU-UCAGGGCCACGCUA-GCAAAA-CCAAUUG-----UUAGCAUAAUGCAAUUAGUGUAAA-AAUCAAA--CAAA .---........(((((((.(((((.(.(((((((((((.....)))-).))))))))((((-((((..-....)))-----)))))....))))).)))))))..-.......--.... ( -36.10) >DroEre_CAF1 798 106 - 1 G----------CUGCACUGCUUGCACGCUGGCCUUAAACUUUGCGUU-UCAGGGUCACGUUA-ACAAAA-CCAAUUGUUAUGUUAGCAUAAUGCAAUUAGUGUAAA-AAUCUAAACUAAA .----------.(((((((.(((((.(.(((((((((((.....)))-).))))))))((((-(((.((-(.....))).)))))))....))))).)))))))..-............. ( -31.80) >DroYak_CAF1 1423 114 - 1 GUUGCACUG--CUGCACUGCUUUCGCGCUGGCCUUAAACUUUGCGUUUUCAGGGUCAUGUUAAACGAAA-CCAAUUGUUAUGUUAGCAUAAUGCAAUUAGCGUAAA-AAUCAAA--CAAA .((((.(((--.(((..((.(((((.(((((((((((((.....))))..))))))).))....)))))-.))...((((((....)))))))))..))).)))).-.......--.... ( -24.00) >consensus A___CACUG__CUGCACUGCUUGCACGCUGGCCUUAAACUUUGCGUU_UCAGGGCCACGUUA_GCAAAA_CCAAUUG_____UUAGCAUAAUGCAAUUAGUGUAAA_AAUCAAA__CAAA ............(((((((.(((((...(((((((.(((.....)))...))))))).((((..(((.......)))......))))....))))).)))))))................ (-18.96 = -19.36 + 0.40)


| Location | 8,337,875 – 8,337,976 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.53 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -16.84 |
| Energy contribution | -17.00 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8337875 101 - 22224390 AACUCAAGCAAGAAGUUAAACGCCAUUUCGGCGAAGCAGUA---CA----------CUGCUUGCACGCUGGCCUUAAACUUUGCGUU-UCAGGGCCACGUUA-GCAAAA-CCAAUUG--- .......((...........((((.....))))((((((..---..----------))))))))(((.(((((((((((.....)))-).))))))))))..-......-.......--- ( -31.50) >DroSec_CAF1 2748 103 - 1 AACUCAAGCAAGAAGUUAAACGCCA---------AGCAGUA---CACUGCACUGCACUGCUUGCACGCUGGCCUUAAACUUUGCGUU-UCAGGGCCACGCUA-GCAAAAAAAAAUUG--- .......((............((.(---------((((((.---((......)).)))))))))..(((((((((((((.....)))-).))))))).))..-))............--- ( -30.20) >DroSim_CAF1 2831 102 - 1 AACUCAAGCAAGAAGUUAAACGCCA---------AGCAGUA---CACUGCACUGCACUGCUUGCACGCUGGCCUUAAACUUUGCGUU-UCAGGGCCACGCUA-GCAAAA-CCAAUUG--- .......((............((.(---------((((((.---((......)).)))))))))..(((((((((((((.....)))-).))))))).))..-))....-.......--- ( -30.20) >DroEre_CAF1 837 107 - 1 AACUCAAGCAAGAAGUUAAACGGCAUGUCGGCUAAGCAGUG----------CUGCACUGCUUGCACGCUGGCCUUAAACUUUGCGUU-UCAGGGUCACGUUA-ACAAAA-CCAAUUGUUA .......(((((.(((....((((((....((...)).)))----------))).))).)))))(((.(((((((((((.....)))-).))))))))))((-((((..-....)))))) ( -30.20) >DroYak_CAF1 1460 117 - 1 AACUCAAGCAAGAAGUUAAACGCCAUGUCGGCUAAGCAGUGUUGCACUG--CUGCACUGCUUUCGCGCUGGCCUUAAACUUUGCGUUUUCAGGGUCAUGUUAAACGAAA-CCAAUUGUUA ......(((((((((((.((.((((((.(((..(((((((((.((...)--).)))))))))))))).)))).)).)))))).(((((.((......))..)))))...-....))))). ( -33.60) >consensus AACUCAAGCAAGAAGUUAAACGCCAU_UCGGC_AAGCAGUA___CACUG__CUGCACUGCUUGCACGCUGGCCUUAAACUUUGCGUU_UCAGGGCCACGUUA_GCAAAA_CCAAUUG___ (((((......).))))..................(((((...............)))))((((..(((((((((.(((.....)))...))))))).))...))))............. (-16.84 = -17.00 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:32 2006