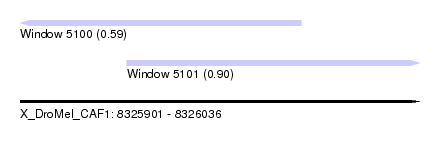
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,325,901 – 8,326,036 |
| Length | 135 |
| Max. P | 0.897875 |

| Location | 8,325,901 – 8,325,996 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.12 |
| Mean single sequence MFE | -15.08 |
| Consensus MFE | 0.00 |
| Energy contribution | 0.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | nan |
| Mean z-score | -1.24 |
| Structure conservation index | -0.00 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8325901 95 - 22224390 AAAU-------AAUUUCAUGCA------------UGAUAAUGCUAUCCAAUAUCUUUCGAAUUGUAUCUCAUAAAAUGCGUACCAUGUUUAUG---CAUAUUUAUAGAAUAGUUAAA ....-------........(((------------(....))))................((((((.(((.((((((((((((.......))))---))).))))))))))))))... ( -14.90) >DroSec_CAF1 19134 94 - 1 AAAU-------AAUUUCAUUCA------------UGAUAAUGCUAUCCAAUAUCUUUCGAAUUGUAUCUCAUAAAUUGCGUACCAUGU-UGUG---CAUACUUAUAGAAUAGUUAAA ...(-------((((..(((((------------(((..((((....((((((....((((((..........)))).))....))))-)).)---)))..)))).))))))))).. ( -11.00) >DroSim_CAF1 19282 94 - 1 AAAU-------AAUUUUAUUCA------------UGAUAAUGCUAUCCCAUAUCUUUCGAAUUGUAUCUCAUAAAUUGCGUACCAUGU-UGUG---CAUAUUUAUAGAAUAGUUAAA ....-------....(((((((------------(((..((((.((..(((......(((.((((.....)))).)))......))).-.)))---)))..)))).))))))..... ( -10.10) >DroEre_CAF1 17695 116 - 1 AAAUUCAUGAUAAUGAUUAUCAUGACAAUGGUUAUCAUACUGAUAACCAAUAUCUUGCAAAUUAUCUGUCAUAAAAUAAGUACCAUGU-CGUGGAUCAUAUCUAUAAAAUAUUUGAA ........((((.(((((..(((((((.((((((((.....)))))))).......(((.......)))................)))-)))))))))))))............... ( -23.60) >DroYak_CAF1 17681 101 - 1 AAAU-------AAUUUCAUUCAU--------UCAUUUCAAUGAUAUCCAAUAUCUUUCACAUUAUCUCUCAUAAAAUGUGUGCCAUGU-UGCGGAUCAUAUCUAUGAAAUAUUUUAA ....-------...........(--------((((....(((((.((((((((....((((((...........))))))....))))-)).)))))))....)))))......... ( -15.80) >consensus AAAU_______AAUUUCAUUCA____________UGAUAAUGCUAUCCAAUAUCUUUCGAAUUGUAUCUCAUAAAAUGCGUACCAUGU_UGUG___CAUAUUUAUAGAAUAGUUAAA ..................................................................................................................... ( 0.00 = 0.00 + 0.00)


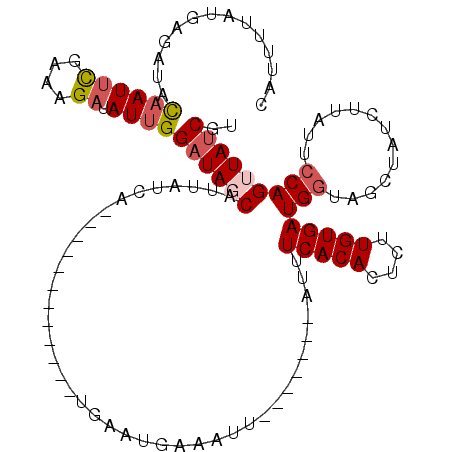
| Location | 8,325,937 – 8,326,036 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.60 |
| Mean single sequence MFE | -23.36 |
| Consensus MFE | -8.54 |
| Energy contribution | -9.66 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.37 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8325937 99 + 22224390 CAUUUUAUGAGAUACAAUUCGAAAGAUAUUGGAUAGCAUUAUCA------------UGCAUGAAAUU-------AUUUCACACUCUUGUGAUGGUAGCUAUCUUAUUCCAGUUAUCGU ..........((((((((((....)).))))((((((..(((((------------(...((((...-------..))))(((....)))))))))))))))..........)))).. ( -21.60) >DroSec_CAF1 19169 99 + 1 CAAUUUAUGAGAUACAAUUCGAAAGAUAUUGGAUAGCAUUAUCA------------UGAAUGAAAUU-------AUUUCACACUCUUGUGAUGGUAGCUAUCUUAUUCCAGUUAUCGU ..........((((((((((....)).))))(((((((((((((------------..(.((((...-------..)))).....)..))))))).))))))..........)))).. ( -22.80) >DroSim_CAF1 19317 99 + 1 CAAUUUAUGAGAUACAAUUCGAAAGAUAUGGGAUAGCAUUAUCA------------UGAAUAAAAUU-------AUUUCACACUCUUGUGAUGGUAGCUAUCUUAUUCCAGUUAUCGU ......((((...((...((....)).(((((((((((((((((------------..(........-------...........)..))))))).))))))))))....))..)))) ( -22.21) >DroEre_CAF1 17733 115 + 1 UAUUUUAUGACAGAUAAUUUGCAAGAUAUUGGUUAUCAGUAUGAUAACCAUUGUCAUGAUAAUCAUUAUCAUGAAUUUCACACUCUUGUGAUGGUAUUU---UUACUGCAGUUAUCAU ............(((((((.(((.((((.((((((((.....)))))))).))))((((((.....))))))((((.((((((....))).))).))))---....)))))))))).. ( -31.10) >DroYak_CAF1 17719 100 + 1 CAUUUUAUGAGAGAUAAUGUGAAAGAUAUUGGAUAUCAUUGAAAUGA--------AUGAAUGAAAUU-------AUUUCACACUCUUGUGAUGGUAUCU---CUACUCCAGUUAUCGU ........((((((((((((.....))))))((((((((((((((((--------.(......).))-------))))))(((....))))))))))))---)).))).......... ( -19.10) >consensus CAUUUUAUGAGAUACAAUUCGAAAGAUAUUGGAUAGCAUUAUCA____________UGAAUGAAAUU_______AUUUCACACUCUUGUGAUGGUAGCUAUCUUAUUCCAGUUAUCGU ..............((((((....)).))))((((((........................................(((((....)))))(((.............))))))))).. ( -8.54 = -9.66 + 1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:24 2006