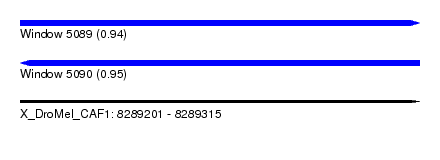
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,289,201 – 8,289,315 |
| Length | 114 |
| Max. P | 0.950764 |

| Location | 8,289,201 – 8,289,315 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 69.83 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -4.19 |
| Energy contribution | -4.83 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.20 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8289201 114 + 22224390 UAUGAAAA-CG----UUGAAAAACGUUCGCUAGAACAUUUCCGCCUUUCGUUACUGGAAAUGCAAUAAUUCCAGAAAUAAUUUUUCCAAAUUGGCUUGCAAUUUUAUUUCAUAACUUUC ((((((((-((----((....)))))..(((((..((((((((...........))))))))...........((((.....))))....)))))...........)))))))...... ( -21.80) >DroSec_CAF1 4925 101 + 1 CAUGAAAU-UG----UUGAAAA--GUUUGCUAGAACAUUUCCGCA-UUCGCUACUGGAAAUGCA---------GCAAUAAU-UUUCCAAAUUCGUUUGCAAUUUUAUUUCAUAAUAUUC .(((((((-((----(.(((((--..(((((....(((((((((.-...))....))))))).)---------))))...)-))))...........))))))))))............ ( -20.10) >DroSim_CAF1 4720 101 + 1 CAUGAAAU-UG----UUGAAAA--GUUUGCUAGAACAUUUCCGCA-UUCGCUGCUGGAAAUGCA---------GCAAUAAU-UUUCGAAAUUGGUUUGCAAUUUUAUUUCAUAAUUUUC .(((((((-..----..(((((--..(((((....((((((((((-.....))).))))))).)---------))))...)-))))(((((((.....))))))))))))))....... ( -26.60) >DroEre_CAF1 4924 103 + 1 UAUGCAAU-CAAGAAUCGAGAU--G-CUGCUAGGAUAUUUCCAUA-UUCGCUACUG-A-AUGUA---------GCAUUACUUUUUCAAAAUUUGUAUGCAAUUUUAUUUCAUAAUUUUC (((((((.-...(((..(.(((--(-((((..(((....)))..(-((((....))-)-)))))---------))))).)...))).....)))))))..................... ( -19.60) >DroYak_CAF1 4656 96 + 1 AAUAUAUUGCAAUAUUCGU--U--A-CUGA-AAUAUAUUGCAAUA-UUCGUUACUG-AAAUGCG---------GCAA----UUUCC--AAUUUCCAUGCAAUUUUAUGACAUAAUUUUC ...(((((((((((((((.--.--.-.)))-...)))))))))))-)..((((..(-((.((((---------(.((----(....--.))).))..))).)))..))))......... ( -17.40) >consensus CAUGAAAU_CG____UUGAAAA__GUUUGCUAGAACAUUUCCGCA_UUCGCUACUGGAAAUGCA_________GCAAUAAUUUUUCCAAAUUGGUUUGCAAUUUUAUUUCAUAAUUUUC .................((((..............((((((((...........))))))))...........((((..................)))).......))))......... ( -4.19 = -4.83 + 0.64)



| Location | 8,289,201 – 8,289,315 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 69.83 |
| Mean single sequence MFE | -21.82 |
| Consensus MFE | -5.22 |
| Energy contribution | -7.26 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.24 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
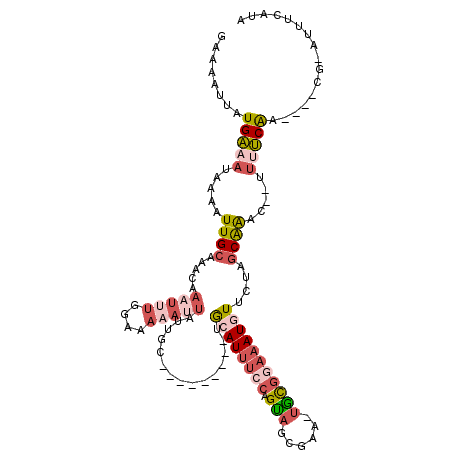
>X_DroMel_CAF1 8289201 114 - 22224390 GAAAGUUAUGAAAUAAAAUUGCAAGCCAAUUUGGAAAAAUUAUUUCUGGAAUUAUUGCAUUUCCAGUAACGAAAGGCGGAAAUGUUCUAGCGAACGUUUUUCAA----CG-UUUUCAUA ......(((((((...........((((((((....)))))....((((((.........))))))........)))((((((((((....))))))))))...----..-.))))))) ( -25.40) >DroSec_CAF1 4925 101 - 1 GAAUAUUAUGAAAUAAAAUUGCAAACGAAUUUGGAAA-AUUAUUGC---------UGCAUUUCCAGUAGCGAA-UGCGGAAAUGUUCUAGCAAAC--UUUUCAA----CA-AUUUCAUG ......((((((((.......((((....))))((((-(...((((---------(((((((((.(((.....-)))))))))))...)))))..--)))))..----..-)))))))) ( -23.20) >DroSim_CAF1 4720 101 - 1 GAAAAUUAUGAAAUAAAAUUGCAAACCAAUUUCGAAA-AUUAUUGC---------UGCAUUUCCAGCAGCGAA-UGCGGAAAUGUUCUAGCAAAC--UUUUCAA----CA-AUUUCAUG ......((((((((.((((((.....)))))).((((-(...((((---------(((((((((.(((.....-)))))))))))...)))))..--)))))..----..-)))))))) ( -26.50) >DroEre_CAF1 4924 103 - 1 GAAAAUUAUGAAAUAAAAUUGCAUACAAAUUUUGAAAAAGUAAUGC---------UACAU-U-CAGUAGCGAA-UAUGGAAAUAUCCUAGCAG-C--AUCUCGAUUCUUG-AUUGCAUA ...................((((..(((...((((........(((---------(((..-.-..))))))..-...(((....)))......-.--...))))...)))-..)))).. ( -15.60) >DroYak_CAF1 4656 96 - 1 GAAAAUUAUGUCAUAAAAUUGCAUGGAAAUU--GGAAA----UUGC---------CGCAUUU-CAGUAACGAA-UAUUGCAAUAUAUU-UCAG-U--A--ACGAAUAUUGCAAUAUAUU ......((((.((......))))))((((((--((...----...)---------)).))))-)........(-(((((((((((...-((..-.--.--..))))))))))))))... ( -18.40) >consensus GAAAAUUAUGAAAUAAAAUUGCAAACAAAUUUGGAAAAAUUAUUGC_________UGCAUUUCCAGUAGCGAA_UGCGGAAAUGUUCUAGCAAAC__UUUUCAA____CG_AUUUCAUA ........(((((.....((((.....(((((....)))))...............((((((((.(((......)))))))))))....)))).....)))))................ ( -5.22 = -7.26 + 2.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:14 2006