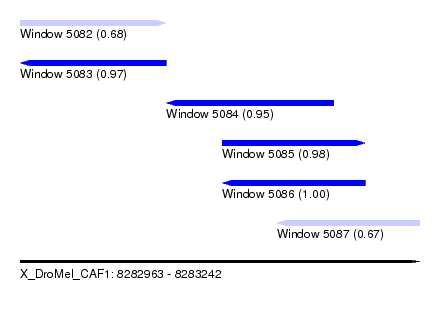
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,282,963 – 8,283,242 |
| Length | 279 |
| Max. P | 0.999900 |

| Location | 8,282,963 – 8,283,065 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.05 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -20.69 |
| Energy contribution | -20.33 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
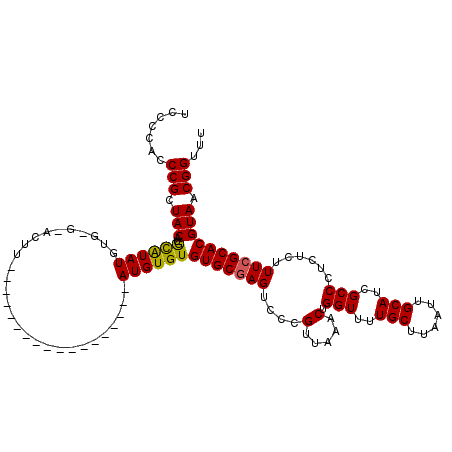
>X_DroMel_CAF1 8282963 102 + 22224390 UCCUCACCCGCUACAUGCAUAUGUGCGUACUU------------------AUGUGUGUGCGAGUCCCGUUAAACUGGUUUUGCUUAAUUGCAUCGCCCUCUCUUUCGCACGUAACGGUUU .......(((.(((..(((((((........)------------------))))))(((((((....(.....).(((..(((......)))..)))......)))))))))).)))... ( -26.70) >DroSec_CAF1 4349 102 + 1 UCCCCACCCGCUACAUGCAUAUGUGUGUACUU------------------AUGUGUGUGCGAGUCCCGUUAAACUGGUUUUGCUUAAUUGCAUCGCCCUCUCUUUCGCACGUAACGGUUU .......(((.(((..(((((((........)------------------))))))(((((((....(.....).(((..(((......)))..)))......)))))))))).)))... ( -26.70) >DroSim_CAF1 4312 102 + 1 UCCCCACCCGCUACAUGCAUAUGUGUGCACUU------------------AUGUGUGUGCUAGUCCCGUUAAACUGGUUUUGCUUAAUUGCAUCGCCCUCUCUUUCGCACGUAACGGUUU .......(((.(((.(((....(((..(((..------------------..)))..)))...............(((..(((......)))..))).........))).))).)))... ( -25.40) >DroEre_CAF1 7471 116 + 1 UCCCCACCCGCUACAUAUGUAUGU----ACUUAGUGUGCGUGUGUGUGUUAUGUGUGUGCGAGUCCCGUUAAACUGGUUUUGCUUAAUGGCAUCGCCCUCUCUUUCGCACGUAACGGUUU .....(((.((.(((((((((..(----.....)..)))))))))))((((((((((.((((...(((((((.(.......).)))))))..)))).........))))))))))))).. ( -36.00) >DroYak_CAF1 3117 98 + 1 UCCCCACCCGCUACAUACAUAUGU----ACUG------------------AUGUGUGUGCGAGUCCCGUUAAACUGGUUUUGCUUAAUUGCAUCGCCCUCUCUUUCGCACGUAACGGUUU .......(((.(((..((((((..----....------------------))))))(((((((....(.....).(((..(((......)))..)))......)))))))))).)))... ( -25.20) >consensus UCCCCACCCGCUACAUGCAUAUGUG_G_ACUU__________________AUGUGUGUGCGAGUCCCGUUAAACUGGUUUUGCUUAAUUGCAUCGCCCUCUCUUUCGCACGUAACGGUUU .......(((.(((..((((((............................))))))(((((((....(.....).(((..(((......)))..)))......)))))))))).)))... (-20.69 = -20.33 + -0.36)


| Location | 8,282,963 – 8,283,065 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.05 |
| Mean single sequence MFE | -29.02 |
| Consensus MFE | -25.28 |
| Energy contribution | -25.28 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8282963 102 - 22224390 AAACCGUUACGUGCGAAAGAGAGGGCGAUGCAAUUAAGCAAAACCAGUUUAACGGGACUCGCACACACAU------------------AAGUACGCACAUAUGCAUGUAGCGGGUGAGGA ...(((((((((((((......((....(((......)))...)).((((....))))))))))......------------------......(((....)))..)))))))....... ( -28.40) >DroSec_CAF1 4349 102 - 1 AAACCGUUACGUGCGAAAGAGAGGGCGAUGCAAUUAAGCAAAACCAGUUUAACGGGACUCGCACACACAU------------------AAGUACACACAUAUGCAUGUAGCGGGUGGGGA ...((((((((((((.........(((((.(..((((((.......))))))..).).))))..((....------------------..)).........))))))))))))....... ( -27.80) >DroSim_CAF1 4312 102 - 1 AAACCGUUACGUGCGAAAGAGAGGGCGAUGCAAUUAAGCAAAACCAGUUUAACGGGACUAGCACACACAU------------------AAGUGCACACAUAUGCAUGUAGCGGGUGGGGA ...((((((((((((.......((....(((......)))...))(((((....))))).((((......------------------..)))).......))))))))))))....... ( -31.00) >DroEre_CAF1 7471 116 - 1 AAACCGUUACGUGCGAAAGAGAGGGCGAUGCCAUUAAGCAAAACCAGUUUAACGGGACUCGCACACACAUAACACACACACGCACACUAAGU----ACAUACAUAUGUAGCGGGUGGGGA ...((((((.((((....).....(((((.((.((((((.......)))))).)).).))))...))).))))...(((.((((((.((.((----....)).))))).))).))))).. ( -29.90) >DroYak_CAF1 3117 98 - 1 AAACCGUUACGUGCGAAAGAGAGGGCGAUGCAAUUAAGCAAAACCAGUUUAACGGGACUCGCACACACAU------------------CAGU----ACAUAUGUAUGUAGCGGGUGGGGA ...(((((((((((((......((....(((......)))...)).((((....))))))))))......------------------..((----((....)))))))))))....... ( -28.00) >consensus AAACCGUUACGUGCGAAAGAGAGGGCGAUGCAAUUAAGCAAAACCAGUUUAACGGGACUCGCACACACAU__________________AAGU_C_CACAUAUGCAUGUAGCGGGUGGGGA ...((((((((((((.........(((((.(..((((((.......))))))..).).))))............................((....))...))))))))))))....... (-25.28 = -25.28 + -0.00)


| Location | 8,283,065 – 8,283,182 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.99 |
| Mean single sequence MFE | -32.74 |
| Consensus MFE | -19.52 |
| Energy contribution | -19.20 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8283065 117 - 22224390 CAGGUAUUUAAAAAAAAUGUCUGUCCAUGUGUGUGCAAGUGCAUGCGUG--UGUGUGGGUGAUGCUGAUAAGCGAAGCUGACGGGAUACGGGUUAAAAGCCGUCAAGAAAUGCCU-AUUU .((((((((..........((((((((..(((((((....)))))))..--))....(((..((((....))))..)))))))))..((((........))))....))))))))-.... ( -36.10) >DroSec_CAF1 4451 103 - 1 CAGGUAUUUGAAAAAAAUGUCUGUGCAUGCGUGU------------G----UGUGUGGGUGAUGCUGAUAAGCAAAGCUGACGGGAUACGGGUUAAAAGCCGUCAAGAAAUGCCU-AUUU (((..((((.....))))..))).((((.(((((------------.----(.(((.(((..((((....))))..))).))).))))))(((.....)))........))))..-.... ( -28.20) >DroSim_CAF1 4414 105 - 1 CAGGUAUUUGAAAAAAAUGUCUGUGCAUGCGUGU------------GUG--UGUGUGGGUGAUGCUGAUAAGCGAAGCUGACGGGAUACGGGUUAAAAGCCGUCAAGAAAUGCCU-AUUU .((((((((.........((((..((((((....------------)))--)))((.(((..((((....))))..))).)).))))((((........))))....))))))))-.... ( -28.10) >DroEre_CAF1 7587 108 - 1 CAGGUAUUUGAAAAAA--AUGUGUGCAUGU--GUGCAAGUGCAUGCGUGUGAGUGUGGGUGAUGCUGAUAAGCGAAGCUGACG-------GGUUAAAAGCCGUCAAGAAAUGCCU-AUUU .((((((((.......--(..(.(((((((--((((....))))))))))).)..).(((..((((....))))..)))((((-------(........)))))...))))))))-.... ( -35.20) >DroYak_CAF1 3215 110 - 1 CAGGUAUUUGAAAAUA-UGUGUGUGCAUUU--GUGCAAGUGCAUGCGUGUGAGUGUGGGUGAUGCUGAUAAGCGAAGUUGACG-------GGUUAAAAGCCGUUAAGAAAUGCCUGAUUU (((((((((.....((-..(((((((((((--....)))))))))))..))...........((((....))))...((((((-------(........))))))).))))))))).... ( -36.10) >consensus CAGGUAUUUGAAAAAAAUGUCUGUGCAUGUGUGUGCAAGUGCAUGCGUG__UGUGUGGGUGAUGCUGAUAAGCGAAGCUGACGGGAUACGGGUUAAAAGCCGUCAAGAAAUGCCU_AUUU .((((((((.............................................((.(((..((((....))))..))).))........(((.....)))......))))))))..... (-19.52 = -19.20 + -0.32)


| Location | 8,283,104 – 8,283,204 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.56 |
| Mean single sequence MFE | -20.34 |
| Consensus MFE | -6.32 |
| Energy contribution | -6.30 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.31 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8283104 100 + 22224390 CAGCUUCGCUUAUCAGCAUCACCCACACA--CACGCAUGCACUUGCACACACAUGGACAGACAUUUUUUUUAAAUACCUGGAAGUGU------------------GCACAUAUGCAGAUA .......(((....)))............--...(((((....(((((((.......(((..((((.....))))..)))...))))------------------)))..)))))..... ( -18.40) >DroSec_CAF1 4490 86 + 1 CAGCUUUGCUUAUCAGCAUCACCCACACA----C------------ACACGCAUGCACAGACAUUUUUUUCAAAUACCUGGUAGUGU------------------GCAUAUAUGCUGAUA ..........(((((((((....((((((----(------------....(....).(((..((((.....))))..))))).))))------------------).....))))))))) ( -18.30) >DroSim_CAF1 4453 88 + 1 CAGCUUCGCUUAUCAGCAUCACCCACACA--CAC------------ACACGCAUGCACAGACAUUUUUUUCAAAUACCUGGUAGUGU------------------GCAUAUAUGCUGAUA ..........(((((((((....((((((--(..------------....(....).(((..((((.....))))..))))).))))------------------).....))))))))) ( -18.30) >DroEre_CAF1 7619 100 + 1 CAGCUUCGCUUAUCAGCAUCACCCACACUCACACGCAUGCACUUGCAC--ACAUGCACACAU--UUUUUUCAAAUACCUGAAAGUGUA--------U--------GUACAUAUGUAGAUA ..........((((.((((...............(((......)))..--((((((((....--...(((((......))))))))))--------)--------))....)))).)))) ( -17.51) >DroYak_CAF1 3248 117 + 1 CAACUUCGCUUAUCAGCAUCACCCACACUCACACGCAUGCACUUGCAC--AAAUGCACACACA-UAUUUUCAAAUACCUGGAAGUGUACAUAUGCAUGUACAUGUGUAUGUAUGUAGAUA .......(((....)))..........((.(((..(((((((.((.((--(..((((.....(-((((((((......))))))))).....))))))).)).)))))))..)))))... ( -29.20) >consensus CAGCUUCGCUUAUCAGCAUCACCCACACA__CACGCAUGCACUUGCACACACAUGCACAGACAUUUUUUUCAAAUACCUGGAAGUGU__________________GCAUAUAUGCAGAUA ..((...(((....))).......((((......(((......))).....................(((((......)))))))))..................))............. ( -6.32 = -6.30 + -0.02)

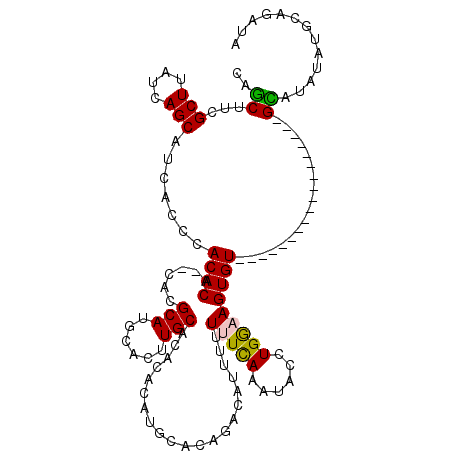
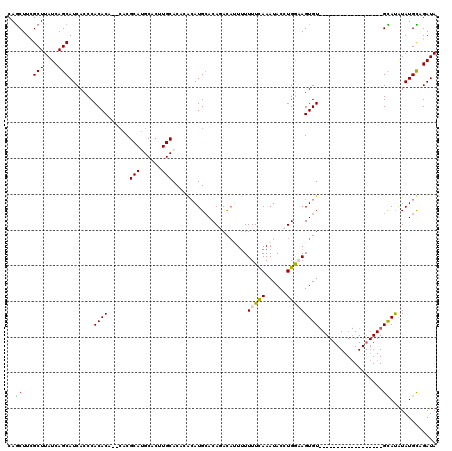
| Location | 8,283,104 – 8,283,204 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.56 |
| Mean single sequence MFE | -33.82 |
| Consensus MFE | -11.14 |
| Energy contribution | -12.48 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.73 |
| Structure conservation index | 0.33 |
| SVM decision value | 4.45 |
| SVM RNA-class probability | 0.999900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8283104 100 - 22224390 UAUCUGCAUAUGUGC------------------ACACUUCCAGGUAUUUAAAAAAAAUGUCUGUCCAUGUGUGUGCAAGUGCAUGCGUG--UGUGUGGGUGAUGCUGAUAAGCGAAGCUG ((((..((((..(((------------------((((...(((..((((.....))))..))).....))))((((....)))))))..--))))..)))).((((....))))...... ( -32.50) >DroSec_CAF1 4490 86 - 1 UAUCAGCAUAUAUGC------------------ACACUACCAGGUAUUUGAAAAAAAUGUCUGUGCAUGCGUGU------------G----UGUGUGGGUGAUGCUGAUAAGCAAAGCUG (((((((((.(((.(------------------((((...(((..((((.....))))..))).((((....))------------)----)))))).))))))))))))(((...))). ( -29.80) >DroSim_CAF1 4453 88 - 1 UAUCAGCAUAUAUGC------------------ACACUACCAGGUAUUUGAAAAAAAUGUCUGUGCAUGCGUGU------------GUG--UGUGUGGGUGAUGCUGAUAAGCGAAGCUG (((((((((.(((.(------------------((((...(((..((((.....))))..))).(((((....)------------)))--)))))).))))))))))))(((...))). ( -30.30) >DroEre_CAF1 7619 100 - 1 UAUCUACAUAUGUAC--------A--------UACACUUUCAGGUAUUUGAAAAAA--AUGUGUGCAUGU--GUGCAAGUGCAUGCGUGUGAGUGUGGGUGAUGCUGAUAAGCGAAGCUG (((((((((((((((--------(--------((...((((((....))))))...--.)))))))))((--((((....))))))......))))))))).((((....))))...... ( -30.20) >DroYak_CAF1 3248 117 - 1 UAUCUACAUACAUACACAUGUACAUGCAUAUGUACACUUCCAGGUAUUUGAAAAUA-UGUGUGUGCAUUU--GUGCAAGUGCAUGCGUGUGAGUGUGGGUGAUGCUGAUAAGCGAAGUUG (((((((((.(((((.(((((((.(((((((((((((....(.(((((....))))-).)))))))))..--))))).))))))).))))).))))))))).((((....))))...... ( -46.30) >consensus UAUCUGCAUAUAUGC__________________ACACUUCCAGGUAUUUGAAAAAAAUGUCUGUGCAUGUGUGUGCAAGUGCAUGCGUG__UGUGUGGGUGAUGCUGAUAAGCGAAGCUG (((((((((...............................(((((((((.....)))))))))..........((((......)))).....))))))))).((((....))))...... (-11.14 = -12.48 + 1.34)

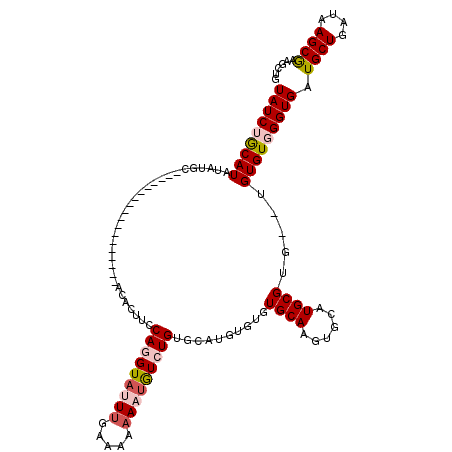
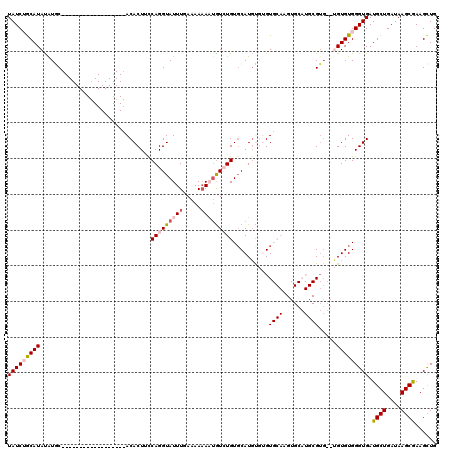
| Location | 8,283,142 – 8,283,242 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.37 |
| Mean single sequence MFE | -22.10 |
| Consensus MFE | -4.88 |
| Energy contribution | -5.40 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.22 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8283142 100 - 22224390 UUAGCCAUUUACU--CUGUGUGCGCUUUGUGUUUAUUAUUUAUCUGCAUAUGUGC------------------ACACUUCCAGGUAUUUAAAAAAAAUGUCUGUCCAUGUGUGUGCAAGU ...(((((.....--..((((((((..(((((..((.....))..))))).))))------------------))))...(((..((((.....))))..))).......))).)).... ( -25.40) >DroSec_CAF1 4520 94 - 1 UUCGCCAUUUACU--CUGUUUGUGCUUUGUGUUUAUUAUUUAUCAGCAUAUAUGC------------------ACACUACCAGGUAUUUGAAAAAAAUGUCUGUGCAUGCGUGU------ ..(((((..(((.--(((..(((((..((((((.((.....)).))))))...))------------------)))....))))))..))......((((....)))))))...------ ( -20.10) >DroSim_CAF1 4485 96 - 1 UUCGCCAUUUACUCUCUGUUUGCGCUUUGUGUUUAUUAUUUAUCAGCAUAUAUGC------------------ACACUACCAGGUAUUUGAAAAAAAUGUCUGUGCAUGCGUGU------ .....................((((..((((((.((.....)).)))))).((((------------------(((......(..((((.....))))..))))))))))))..------ ( -17.20) >DroEre_CAF1 7659 96 - 1 UUCGCUAUUUACU--CAAU--GUGCUUUGUGUUUAUUAUUUAUCUACAUAUGUAC--------A--------UACACUUUCAGGUAUUUGAAAAAA--AUGUGUGCAUGU--GUGCAAGU ...((.....((.--(((.--.....))).)).............((((((((((--------(--------((...((((((....))))))...--.)))))))))))--)))).... ( -20.70) >DroYak_CAF1 3288 113 - 1 UUCGCCAUUUACU--CAGU--GUGCUUUGUGUUUAUUGUUUAUCUACAUACAUACACAUGUACAUGCAUAUGUACACUUCCAGGUAUUUGAAAAUA-UGUGUGUGCAUUU--GUGCAAGU ...((........--..((--((((..(((((....(((......))).....))))).))))))(((.((((((((....(.(((((....))))-).))))))))).)--)))).... ( -27.10) >consensus UUCGCCAUUUACU__CUGU_UGUGCUUUGUGUUUAUUAUUUAUCUGCAUAUAUGC__________________ACACUUCCAGGUAUUUGAAAAAAAUGUCUGUGCAUGUGUGUGCAAGU ...((................(((...(((((..((.....))..)))))..))).........................(((((((((.....))))))))).)).............. ( -4.88 = -5.40 + 0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:11 2006