
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,281,478 – 8,281,612 |
| Length | 134 |
| Max. P | 0.639773 |

| Location | 8,281,478 – 8,281,578 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 80.59 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -23.79 |
| Energy contribution | -23.93 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8281478 100 - 22224390 GCGCGUCAUGCCGGCGUCCUGGAGCAGCGGGGGCAGCCUGCUGCCCUUUCGCGUUUCCACCACCACAAAGGUGAGCCUAGCUAACUUAAGUA----CCCAGAAC ..(((((.....))))).(((((((.((((((((((....)))))))..)))........((((.....))))......))).((....)).----.))))... ( -35.00) >DroSec_CAF1 2860 100 - 1 GCGCGUCAUGCCGGCAUCUUGGAGCAGCGGGGGCAGUCUGCUGCCGUUUCGCGUUUCCACCACCACAAAGGUGAGCUUAGCUAACUUAGAAA----UACAGACU ....(((..((.(((....(((((..((((((((((....)))))..)))))..))))).((((.....)))).)))..)).....((....----))..))). ( -30.30) >DroSim_CAF1 2824 100 - 1 GCGCGUCAUGCCGGCAUCUUGGAGCAGCGGGGGCAGUCUGCUGCCAUUUCGCGUUUCCACCACCACAAAGGUGAGCUUAGCUAACUUAAGUA----GCCAGACU ....(((.....(((....(((((..((((.(((((....)))))...))))..))))).((((.....)))).)))..((((.(....)))----))..))). ( -32.70) >DroEre_CAF1 5992 100 - 1 CCGUGUCAUGCCGGCAUCCUGGACAAGCGGGGGCAGCCUGCUGCCCUUCCGCGUUUCCACCACCACAAAGGUGGGAUAAUUUACCAUAAGUA----CACAGACU ..(((((.((((((....)))).)).((((((((((....))))))..))))..(((((((........))))))).............).)----)))..... ( -37.30) >DroYak_CAF1 1597 104 - 1 GCGUGUCAUGCCGGCAUCCUGGACUAGCGGGGGCAGCCUGCUGCCCUUUCGCGUUUCCACCACCACAAAGGUGGGCUUAUUUACCUAAAGUAUUAAUAAAGACU ....(((.....(((....((((...((((((((((....)))))))..)))...))))(((((.....))))))))(((((.....)))))........))). ( -35.50) >DroAna_CAF1 2916 100 - 1 GCGCAUUAUGCCCGCCACCUGGACCAGUGGUGGCGGCCUCCUGCCCUUUCGCAUCUCCACCACCACAAAGGUAGGCCUAGAUAGACUCAUAU----CCUAGACC (.((.....)).)(((((((......(((((((.((.....(((......)))...)).)))))))..)))).)))(((((((......)))----.))))... ( -30.10) >consensus GCGCGUCAUGCCGGCAUCCUGGACCAGCGGGGGCAGCCUGCUGCCCUUUCGCGUUUCCACCACCACAAAGGUGAGCUUAGCUAACUUAAGUA____CACAGACU ............(((....((((...((((((((((....)))))..)))))...)))).((((.....)))).)))........................... (-23.79 = -23.93 + 0.14)


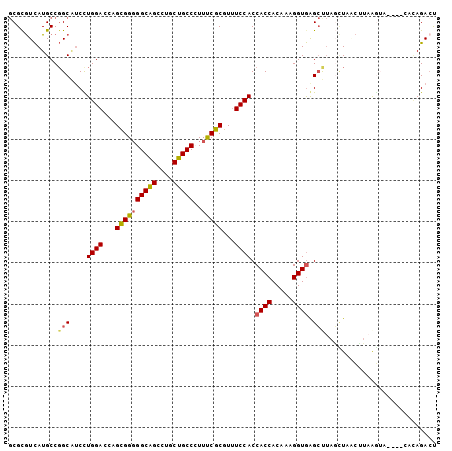
| Location | 8,281,501 – 8,281,612 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 83.19 |
| Mean single sequence MFE | -45.68 |
| Consensus MFE | -33.39 |
| Energy contribution | -33.12 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8281501 111 - 22224390 -----AUCCAAG-GCGUAGUCAUCAUAGCCGCAGGAUGCAGCGCGUCAUGCCGGCGUCCUGGAGCAGCGGGGGCAGCCUGCUGCCCUUUCGCGUUUCCACCACCACAAAGGUGAGCC -----......(-((............((((((.(((((...))))).)).))))....(((((..((((((((((....)))))))..)))..))))).((((.....)))).))) ( -45.60) >DroSec_CAF1 2883 111 - 1 -----AUCCAAG-GCGUAGUCAUCGUAGCCGCAGGAUGCAGCGCGUCAUGCCGGCAUCUUGGAGCAGCGGGGGCAGUCUGCUGCCGUUUCGCGUUUCCACCACCACAAAGGUGAGCU -----.((((((-(((.......))).((((((.(((((...))))).)).))))..))))))((.((((((((((....)))))..)))))........((((.....)))).)). ( -44.70) >DroSim_CAF1 2847 111 - 1 -----AUCCAAG-GCGUAGUCAUCGUAGCCGCAGGAUGCAGCGCGUCAUGCCGGCAUCUUGGAGCAGCGGGGGCAGUCUGCUGCCAUUUCGCGUUUCCACCACCACAAAGGUGAGCU -----.((((((-(((.......))).((((((.(((((...))))).)).))))..))))))((.((((.(((((....)))))...))))........((((.....)))).)). ( -45.20) >DroEre_CAF1 6015 111 - 1 -----AUCCACG-GCGGAAUCAUACUAACCGCAGGAUGCACCGUGUCAUGCCGGCAUCCUGGACAAGCGGGGGCAGCCUGCUGCCCUUCCGCGUUUCCACCACCACAAAGGUGGGAU -----.(((...-((((...........))))(((((((.(.((.....)).)))))))))))...((((((((((....))))))..))))..(((((((........))))))). ( -49.10) >DroYak_CAF1 1624 111 - 1 -----AUCCACA-GCGGAAUCAUUUUAGCCGCAGGAUGCAGCGUGUCAUGCCGGCAUCCUGGACUAGCGGGGGCAGCCUGCUGCCCUUUCGCGUUUCCACCACCACAAAGGUGGGCU -----......(-((((((.(....(((.(.((((((((.((((...))))..))))))))).)))((((((((((....)))))))..)))).)))).(((((.....)))))))) ( -51.40) >DroAna_CAF1 2939 113 - 1 AAGUGUUCCAAGAGUGAGGUGACCA----CGAAGGAUGCAGCGCAUUAUGCCCGCCACCUGGACCAGUGGUGGCGGCCUCCUGCCCUUUCGCAUCUCCACCACCACAAAGGUAGGCC .............(.((((((....----((((((..((((.((.....))((((((((.........))))))))....)))).))))))))))))).(((((.....))).)).. ( -38.10) >consensus _____AUCCAAG_GCGUAGUCAUCAUAGCCGCAGGAUGCAGCGCGUCAUGCCGGCAUCCUGGACCAGCGGGGGCAGCCUGCUGCCCUUUCGCGUUUCCACCACCACAAAGGUGAGCU .............(((.............)))(((((((...((.....))..)))))))(((...((((((((((....)))))..)))))...)))..((((.....)))).... (-33.39 = -33.12 + -0.27)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:03 2006