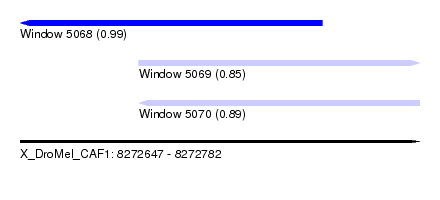
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,272,647 – 8,272,782 |
| Length | 135 |
| Max. P | 0.989449 |

| Location | 8,272,647 – 8,272,749 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 63.24 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -19.68 |
| Energy contribution | -19.93 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8272647 102 - 22224390 CAUUGCCAUUGCUGGCGACGGUCGGCAGA-UUAGCCUUAUUUUC-----------------GCUGCUAGUAAGGAACAUAUUUUUACUGCCGGUUGCCAGAACAAUGGUAUCCCCGUUGG ...((((((((((((((((...((((((.-....(((((((...-----------------......)))))))....((....))))))))))))))))..)))))))).......... ( -35.80) >DroSim_CAF1 11872 102 - 1 CAUUGCCAUUGCUGGCGACGGUCGGCAGA-AUAGCCUUAUUAUC-----------------GCUGCUAGUAAGGAACAUACUUUUGCUGCCGGUUGCCAGCACAAUGGUAUCCCCGUUGG ...((((((((((((((((((.(((((((-(((.((((((((..-----------------.....))))))))....)).)))))))))).))))))))))..)))))).......... ( -43.20) >DroWil_CAF1 16489 111 - 1 GAUUACCACCAUUGAGAAUAGUCGUUGUUCCUGUCCCCGUUACAGCUGCUGCUUGCUUCUCGGCAUUGGAAAAUAUCAUUGGUUUAUUGCCAUUUGCCAGGACA---------CCGUUGA ........((...(((((((((.(((((..(.......)..))))).)))).....)))))((((.(((..((((.........)))).)))..)))).))...---------....... ( -23.10) >DroYak_CAF1 10059 105 - 1 CAUUGCCAUUGGUGGCGACGGUCGGCAUC-ACACCCUUGUUAUCAUU--------------ACUGCUGGUAAGGAACAUACUCUUGUUGCCGUUCGCCAGCACAAUGGUAUCGCCGUUGG ..((((((....))))))(((.((((...-(((....)))(((((((--------------..(((((((..((((((......)))).))....))))))).)))))))..))))))). ( -35.10) >consensus CAUUGCCAUUGCUGGCGACGGUCGGCAGA_AUAGCCUUAUUAUC_________________GCUGCUAGUAAGGAACAUACUUUUACUGCCGGUUGCCAGCACAAUGGUAUCCCCGUUGG .........(((((((((((((.(((((..................................)))))(((((((.......))))))))))).)))))))))((((((.....)))))). (-19.68 = -19.93 + 0.25)



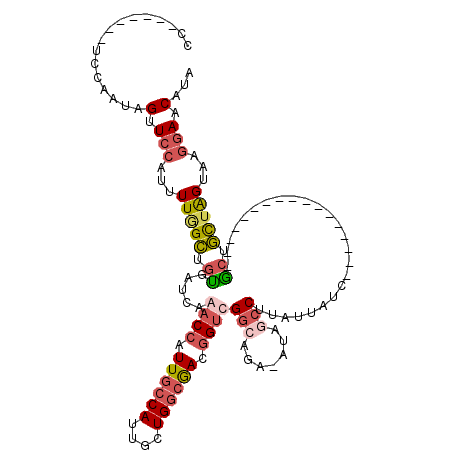
| Location | 8,272,687 – 8,272,782 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 59.60 |
| Mean single sequence MFE | -29.08 |
| Consensus MFE | -13.90 |
| Energy contribution | -15.90 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8272687 95 + 22224390 UAUGUUCCUUACUAGCAGC-----------------GAAAAUAAGGCUAA-UCUGCCGACCGUCGCCAGCAAUGGCAAUGGUCUGAUCACAGCCAAAAUGGAUCUAUUGGA-------GG ....((((...........-----------------........(((...-...)))((((((.((((....)))).)))))).((((.((.......))))))....)))-------). ( -29.00) >DroSim_CAF1 11912 95 + 1 UAUGUUCCUUACUAGCAGC-----------------GAUAAUAAGGCUAU-UCUGCCGACCGUCGCCAGCAAUGGCAAUGGUCUGAUCACGGCCAAAAUGGAGCUAUUGGA-------GG ...(((((......((.(.-----------------(((.....(((...-...)))((((((.((((....)))).))))))..))).).))......))))).......-------.. ( -33.00) >DroWil_CAF1 16520 120 + 1 AAUGAUAUUUUCCAAUGCCGAGAAGCAAGCAGCAGCUGUAACGGGGACAGGAACAACGACUAUUCUCAAUGGUGGUAAUCGAUUUAUGGUUACAAAAAUGGAACAUCCAGAUACAACAGC .......(((((.......)))))((.....)).(((((...(....).(((...((.(((((.....))))).))..((.((((.((....)).)))).))...))).......))))) ( -22.20) >DroYak_CAF1 10099 98 + 1 UAUGUUCCUUACCAGCAGU--------------AAUGAUAACAAGGGUGU-GAUGCCGACCGUCGCCACCAAUGGCAAUGGUUUGCUCACAGCUAAAACGGAACUAUUGGA-------GG ...(((((.......((..--------------..)).......((.(((-((.((.((((((.((((....)))).)))))).))))))).)).....))))).......-------.. ( -32.10) >consensus UAUGUUCCUUACCAGCAGC_________________GAUAACAAGGCUAU_UAUGCCGACCGUCGCCAGCAAUGGCAAUGGUCUGAUCACAGCCAAAAUGGAACUAUUGGA_______GG ...(((((......((............................(((.......)))((((((.((((....)))).))))))........))......)))))................ (-13.90 = -15.90 + 2.00)

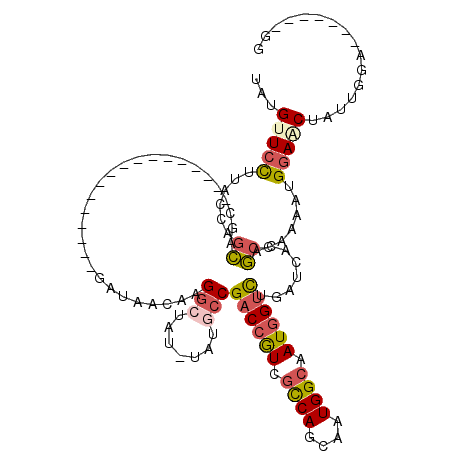
| Location | 8,272,687 – 8,272,782 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 59.60 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -13.18 |
| Energy contribution | -14.92 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8272687 95 - 22224390 CC-------UCCAAUAGAUCCAUUUUGGCUGUGAUCAGACCAUUGCCAUUGCUGGCGACGGUCGGCAGA-UUAGCCUUAUUUUC-----------------GCUGCUAGUAAGGAACAUA ..-------.......(.(((...(((((.((((...((((.((((((....)))))).))))(((...-...)))......))-----------------)).)))))...))).)... ( -32.40) >DroSim_CAF1 11912 95 - 1 CC-------UCCAAUAGCUCCAUUUUGGCCGUGAUCAGACCAUUGCCAUUGCUGGCGACGGUCGGCAGA-AUAGCCUUAUUAUC-----------------GCUGCUAGUAAGGAACAUA ..-------.......(.(((...(((((.(((((..((((.((((((....)))))).))))(((...-...))).....)))-----------------)).)))))...))).)... ( -33.00) >DroWil_CAF1 16520 120 - 1 GCUGUUGUAUCUGGAUGUUCCAUUUUUGUAACCAUAAAUCGAUUACCACCAUUGAGAAUAGUCGUUGUUCCUGUCCCCGUUACAGCUGCUGCUUGCUUCUCGGCAUUGGAAAAUAUCAUU .............(((((((((.....((((.(.......).)))).....(((((((((((.(((((..(.......)..))))).)))).....)))))))...))))..)))))... ( -19.40) >DroYak_CAF1 10099 98 - 1 CC-------UCCAAUAGUUCCGUUUUAGCUGUGAGCAAACCAUUGCCAUUGGUGGCGACGGUCGGCAUC-ACACCCUUGUUAUCAUU--------------ACUGCUGGUAAGGAACAUA ..-------.......(((((...((((((((((((..(((.((((((....)))))).)))..)).))-)))....((....))..--------------...)))))...)))))... ( -32.60) >consensus CC_______UCCAAUAGUUCCAUUUUGGCUGUGAUCAAACCAUUGCCAUUGCUGGCGACGGUCGGCAGA_AUAGCCUUAUUAUC_________________GCUGCUAGUAAGGAACAUA ................(.(((...(((((.((.....((((.((((((....)))))).))))(((.......))).........................)).)))))...))).)... (-13.18 = -14.92 + 1.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:57 2006