
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,258,853 – 8,259,029 |
| Length | 176 |
| Max. P | 0.838703 |

| Location | 8,258,853 – 8,258,949 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -21.53 |
| Consensus MFE | -17.69 |
| Energy contribution | -17.37 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8258853 96 + 22224390 CUCACUUUGCUUGUGUUUGUUGGUUUCUGCUGGAAAACGAGCAUAAGCAGUUAAUAAUUGGCAGUGAUUGCAAUAAAUGCAAAUAUGUAUUCUUUU .(((((.((((((((((((((..((((....))))))))))))))))))((((.....)))))))))(((((.....))))).............. ( -27.40) >DroSec_CAF1 9115 89 + 1 CUCAGUUUGCUUUUGUU---UGGUUUCUGCUGGAAAACGAGCAUAAGCAGUUAAUAAUUGGCAGUGAUUGCAAUAAAUGCAAAU----GUUCUUUU ....((((((.((((((---(((((.((((..(..(((..((....)).))).....)..)))).))))).)))))).))))))----........ ( -19.90) >DroYak_CAF1 13714 89 + 1 UUCACUUUGCUUUUGUU---UGGUUUUUGCUGGAAAACGGGCAUAAACAGUUAAUAAUUGGCUAUGAUUGCAAUAAAUGCAAAU----AUUCUUUU .(((....(((.(((((---..((((.((((.(....).)))).))))....)))))..)))..)))(((((.....)))))..----........ ( -17.30) >consensus CUCACUUUGCUUUUGUU___UGGUUUCUGCUGGAAAACGAGCAUAAGCAGUUAAUAAUUGGCAGUGAUUGCAAUAAAUGCAAAU____AUUCUUUU .(((..(((((..((((.....((((.((((.(....).)))).))))....))))...))))))))(((((.....))))).............. (-17.69 = -17.37 + -0.33)



| Location | 8,258,869 – 8,258,989 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.10 |
| Mean single sequence MFE | -25.81 |
| Consensus MFE | -18.66 |
| Energy contribution | -18.00 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8258869 120 + 22224390 UUGUUGGUUUCUGCUGGAAAACGAGCAUAAGCAGUUAAUAAUUGGCAGUGAUUGCAAUAAAUGCAAAUAUGUAUUCUUUUUUAAAUGGAGCAUGAAAAAAGAAUAUAAGGGGGAAAAUCA ((((..(((.(((((..............))))).....)))..))))((((((((.....))).....(((((((((((((..(((...))).)))))))))))))........))))) ( -25.04) >DroSec_CAF1 9131 113 + 1 U---UGGUUUCUGCUGGAAAACGAGCAUAAGCAGUUAAUAAUUGGCAGUGAUUGCAAUAAAUGCAAAU----GUUCUUUUUUAAAUGGAGCAUGCAAAAAGAAUAUAAGGGGAGAAAUCA .---((((((((.((.........((((..(((((((...........))))))).....))))..((----(((((((((...(((...)))..)))))))))))...)).)))))))) ( -25.60) >DroYak_CAF1 13730 110 + 1 U---UGGUUUUUGCUGGAAAACGGGCAUAAACAGUUAAUAAUUGGCUAUGAUUGCAAUAAAUGCAAAU----AUUCUUUUUUAAAUGGAGCAUGUAAAAAGAAUAUAGGG---GAAACCA .---..((((.((((.(....).)))).))))(((((.....)))))....(((((.....)))))((----(((((((((...(((...)))..)))))))))))..((---....)). ( -26.80) >consensus U___UGGUUUCUGCUGGAAAACGAGCAUAAGCAGUUAAUAAUUGGCAGUGAUUGCAAUAAAUGCAAAU____AUUCUUUUUUAAAUGGAGCAUGAAAAAAGAAUAUAAGGGG_GAAAUCA ......((((.((((.(....).)))).)))).((((.....)))).....(((((.....)))))......(((((((((...(((...)))..)))))))))................ (-18.66 = -18.00 + -0.66)

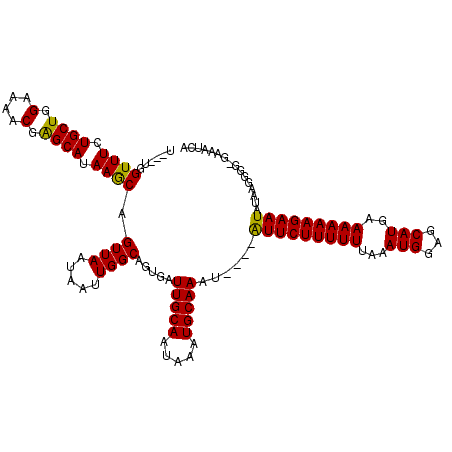

| Location | 8,258,869 – 8,258,989 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.10 |
| Mean single sequence MFE | -16.51 |
| Consensus MFE | -11.94 |
| Energy contribution | -11.39 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8258869 120 - 22224390 UGAUUUUCCCCCUUAUAUUCUUUUUUCAUGCUCCAUUUAAAAAAGAAUACAUAUUUGCAUUUAUUGCAAUCACUGCCAAUUAUUAACUGCUUAUGCUCGUUUUCCAGCAGAAACCAACAA (((............(((((((((((.(........).))))))))))).....(((((.....))))))))((((........(((.((....))..))).....)))).......... ( -17.12) >DroSec_CAF1 9131 113 - 1 UGAUUUCUCCCCUUAUAUUCUUUUUGCAUGCUCCAUUUAAAAAAGAAC----AUUUGCAUUUAUUGCAAUCACUGCCAAUUAUUAACUGCUUAUGCUCGUUUUCCAGCAGAAACCA---A (((..............((((((((..(((...)))...)))))))).----..(((((.....))))))))((((........(((.((....))..))).....))))......---. ( -14.92) >DroYak_CAF1 13730 110 - 1 UGGUUUC---CCCUAUAUUCUUUUUACAUGCUCCAUUUAAAAAAGAAU----AUUUGCAUUUAUUGCAAUCAUAGCCAAUUAUUAACUGUUUAUGCCCGUUUUCCAGCAAAAACCA---A (((((..---....(((((((((((..(((...)))...)))))))))----))(((((.....)))))....)))))..........((((.(((..........))).))))..---. ( -17.50) >consensus UGAUUUC_CCCCUUAUAUUCUUUUUACAUGCUCCAUUUAAAAAAGAAU____AUUUGCAUUUAUUGCAAUCACUGCCAAUUAUUAACUGCUUAUGCUCGUUUUCCAGCAGAAACCA___A (((((............((((((((..(((...)))...)))))))).........(((.....))))))))((((........(((.((....))..))).....)))).......... (-11.94 = -11.39 + -0.55)


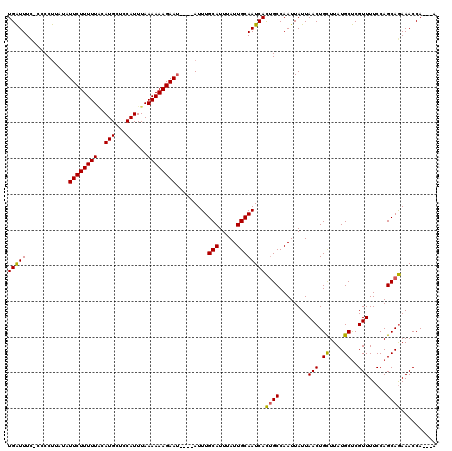
| Location | 8,258,909 – 8,259,029 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.33 |
| Mean single sequence MFE | -24.12 |
| Consensus MFE | -17.37 |
| Energy contribution | -16.93 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8258909 120 + 22224390 AUUGGCAGUGAUUGCAAUAAAUGCAAAUAUGUAUUCUUUUUUAAAUGGAGCAUGAAAAAAGAAUAUAAGGGGGAAAAUCAGCAGAGAUUGCAAAUAAAAACUAUAAACAAACCACGGAGU .......(((.(((((((...(((.....(((((((((((((..(((...))).)))))))))))))..((......)).)))...)))))))(((.....)))........)))..... ( -21.80) >DroSec_CAF1 9168 116 + 1 AUUGGCAGUGAUUGCAAUAAAUGCAAAU----GUUCUUUUUUAAAUGGAGCAUGCAAAAAGAAUAUAAGGGGAGAAAUCAGCAGAGAUUGCAAAUAAAAACUAUAAACAAACCACGAAAU .......(((.(((((((...(((..((----(((((((((...(((...)))..))))))))))).....((....)).)))...)))))))(((.....)))........)))..... ( -23.00) >DroYak_CAF1 13767 113 + 1 AUUGGCUAUGAUUGCAAUAAAUGCAAAU----AUUCUUUUUUAAAUGGAGCAUGUAAAAAGAAUAUAGGG---GAAACCAGCAGAGAUUGCAAAAAAAAACUAUAAACAAAGCACGAAGU .((((((.((.(((((((...(((..((----(((((((((...(((...)))..)))))))))))..((---....)).)))...)))))))..............)).))).)))... ( -27.56) >consensus AUUGGCAGUGAUUGCAAUAAAUGCAAAU____AUUCUUUUUUAAAUGGAGCAUGAAAAAAGAAUAUAAGGGG_GAAAUCAGCAGAGAUUGCAAAUAAAAACUAUAAACAAACCACGAAGU ...........(((((((...(((........(((((((((...(((...)))..)))))))))....((.......)).)))...)))))))........................... (-17.37 = -16.93 + -0.44)


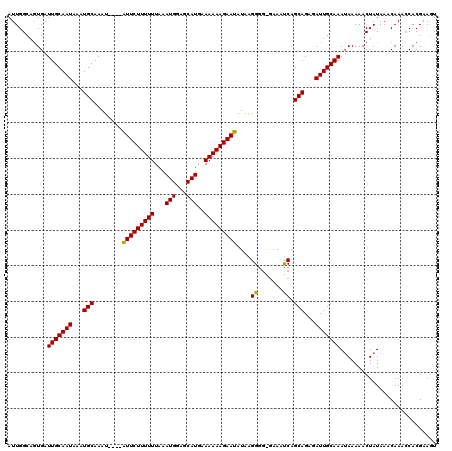
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:50 2006