| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,241,814 – 8,241,917 |
| Length | 103 |
| Max. P | 0.705322 |

| Location | 8,241,814 – 8,241,917 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.54 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -18.17 |
| Energy contribution | -18.84 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8241814 103 + 22224390 -GUAUGGGUAUGGGUUUUUUUUUUGGUAGGGUUUU--------------CCACACA--GCCAAGCAAAUCCCGUGGCAACUUUUUGCCACGCGAAAACUUUCUCAGGCAAGUUUAUGCAU -..........(((...(((.((((((..((....--------------)).....--)))))).))).)))(((((((....)))))))(((.((((((.(....).)))))).))).. ( -29.50) >DroSec_CAF1 142870 93 + 1 -GCAUGGG----------UUUUUCGGUAGGGUUUU--------------CCACACA--GCCAAGCAAAUCCCGUGGCAACUUUUUGCCACGCGAAAACUUUCUCAGGCAAGUUUAUGCAU -(((((..----------(((.(((..((((((((--------------(......--((...))......((((((((....)))))))).)))))))))..).)).)))..))))).. ( -28.50) >DroSim_CAF1 106607 91 + 1 ---AUGGG----------UUUUUCGGUAGGGUUUU--------------CCACACA--GCCAAGCAAAUCCCGUGGCAACUUUUUGCCACGCGAAAACUUUCUCAGGCAAGUUUAUGCAU ---..(((----------.......((..((((..--------------......)--)))..))....)))(((((((....)))))))(((.((((((.(....).)))))).))).. ( -26.20) >DroEre_CAF1 131474 91 + 1 -G-UUGGG----G-----U-UUUCGGUUGGGUUUU--------------CCAC-CA--GCCAAGCAAAUACCGUGGCAACUUUUUGCCACGCGAAAACUUUCUCAGGCAAGUUUAUGCAU -(-..(((----(-----(-((((((((((.....--------------...)-))--)))..........((((((((....)))))))).)))))))))..)..(((......))).. ( -33.10) >DroYak_CAF1 132432 110 + 1 GG-UUGGG----GG----UUUUUCGGUAGGGUUUUCACUAGGGUUUUCCCCAC-CAUGGCCAAGCAAAUCCCGUGGCAACUUUUUGCCACGCGAAAACUUUCUCAGGCAAGUUUAUGCAU ..-...((----((----(((((.(((..(((........(((.....)))))-)...))).)).)))))))(((((((....)))))))(((.((((((.(....).)))))).))).. ( -35.10) >DroAna_CAF1 208893 89 + 1 -GUAGGGG----------UUGGGGGGUGGGGAGGU--------------ACA---A--CCCAAGCAAAUCCCAUGGCAACUUUUUGCCACGCGA-AACUUUCUCAGGCAAGUUUAUGCAU -...((((----------((...(..((((.....--------------...---.--))))..).)))))).((((((....)))))).((((-(((((.(....).)))))).))).. ( -28.40) >consensus _G_AUGGG__________UUUUUCGGUAGGGUUUU______________CCAC_CA__GCCAAGCAAAUCCCGUGGCAACUUUUUGCCACGCGAAAACUUUCUCAGGCAAGUUUAUGCAU .....(((.................((..(((..........................)))..))....)))(((((((....)))))))(((.((((((.(....).)))))).))).. (-18.17 = -18.84 + 0.67)



| Location | 8,241,814 – 8,241,917 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.54 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -19.70 |
| Energy contribution | -19.73 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8241814 103 - 22224390 AUGCAUAAACUUGCCUGAGAAAGUUUUCGCGUGGCAAAAAGUUGCCACGGGAUUUGCUUGGC--UGUGUGG--------------AAAACCCUACCAAAAAAAAAACCCAUACCCAUAC- ..(((.((((((........))))))((.((((((((....)))))))).))..))).(((.--.((((((--------------......................)))))))))...- ( -27.55) >DroSec_CAF1 142870 93 - 1 AUGCAUAAACUUGCCUGAGAAAGUUUUCGCGUGGCAAAAAGUUGCCACGGGAUUUGCUUGGC--UGUGUGG--------------AAAACCCUACCGAAAAA----------CCCAUGC- ..((..((((((........))))))..))(((((((....)))))))(((.(((..((((.--.(.((..--------------...)).)..)))).)))----------)))....- ( -27.00) >DroSim_CAF1 106607 91 - 1 AUGCAUAAACUUGCCUGAGAAAGUUUUCGCGUGGCAAAAAGUUGCCACGGGAUUUGCUUGGC--UGUGUGG--------------AAAACCCUACCGAAAAA----------CCCAU--- ..((..((((((........))))))..))(((((((....)))))))(((.(((..((((.--.(.((..--------------...)).)..)))).)))----------)))..--- ( -27.00) >DroEre_CAF1 131474 91 - 1 AUGCAUAAACUUGCCUGAGAAAGUUUUCGCGUGGCAAAAAGUUGCCACGGUAUUUGCUUGGC--UG-GUGG--------------AAAACCCAACCGAAA-A-----C----CCCAA-C- ..((..((((((........))))))..))(((((((....)))))))((..(((..((((.--((-(.(.--------------....)))).))))))-)-----.----.))..-.- ( -26.40) >DroYak_CAF1 132432 110 - 1 AUGCAUAAACUUGCCUGAGAAAGUUUUCGCGUGGCAAAAAGUUGCCACGGGAUUUGCUUGGCCAUG-GUGGGGAAAACCCUAGUGAAAACCCUACCGAAAAA----CC----CCCAA-CC ............(((.....((((..((.((((((((....)))))))).))...)))))))..((-((((((...((....)).....)))))))).....----..----.....-.. ( -33.30) >DroAna_CAF1 208893 89 - 1 AUGCAUAAACUUGCCUGAGAAAGUU-UCGCGUGGCAAAAAGUUGCCAUGGGAUUUGCUUGGG--U---UGU--------------ACCUCCCCACCCCCCAA----------CCCCUAC- ..(((......)))......((((.-((.((((((((....)))))))).))...))))(((--(---((.--------------..............)))----------)))....- ( -25.36) >consensus AUGCAUAAACUUGCCUGAGAAAGUUUUCGCGUGGCAAAAAGUUGCCACGGGAUUUGCUUGGC__UG_GUGG______________AAAACCCUACCGAAAAA__________CCCAU_C_ ..(((......)))......((((..((.((((((((....)))))))).))...))))............................................................. (-19.70 = -19.73 + 0.03)

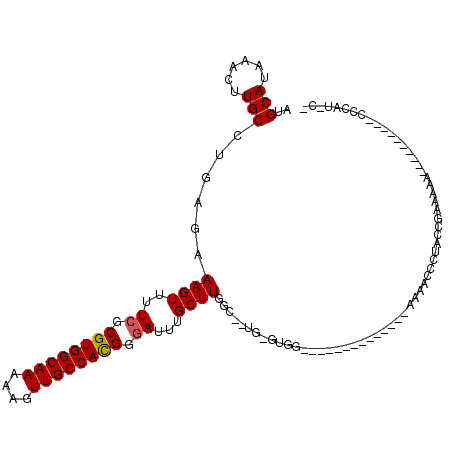

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:38 2006