
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,231,921 – 8,232,085 |
| Length | 164 |
| Max. P | 0.993205 |

| Location | 8,231,921 – 8,232,022 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 88.12 |
| Mean single sequence MFE | -38.43 |
| Consensus MFE | -28.73 |
| Energy contribution | -30.54 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8231921 101 + 22224390 UCCCGCACCGGAUUCCUGGUCGGAUCCUUCCGCAAAAGCAAAGCGAAACGGGAGCGCCAGAGCCGAUCGAUGCGCAAAAGCAAGGACUCCAAAGAGGAGUC (((((((((((....))))).(((....)))))....((...)).....))))((((..((.....))...)))).........((((((.....)))))) ( -33.20) >DroSec_CAF1 132824 101 + 1 UCCCGCACCGAGUUCCUGGUCGGAUCCUUGCGCAAAAACAAAGCGAAGCGGGAGCGCCAGAGCCGCUCGAUGCGCAAAAGCAGGGACUCCAAAGAGGAGUC (((((((.(((((.(((((.((..((((..(((.........)))....)))).)))))).)..))))).)))((....)).))))((((.....)))).. ( -38.70) >DroEre_CAF1 121934 101 + 1 UCCCGCACCGAGAUCCUGGUCGGAUCCUUGCGCAAGAGCAAGGCGAAGCGGGAGCGCAAAAGCCGUUCGAUGCGCAAGUGCAUGGACUCCAAAGAGGAGUC ....((((...(((((.....))))).(((((((.((((..(((...(((....)))....)))))))..)))))))))))...((((((.....)))))) ( -44.20) >DroYak_CAF1 122994 101 + 1 UCCCGCACCGAGAUCCUGGUCGGUUCCUUGCGCAAGAGCAAGGCGAGGCGGGAGCGCAAAAGUCGUUCGAUGCGCAAAAGCAGCGACUCCAAAGAGGACAA ((((((..((.(((....)))....((((((......))))))))..))))))........(((..(((.(((......))).)))(((....)))))).. ( -37.60) >consensus UCCCGCACCGAGAUCCUGGUCGGAUCCUUGCGCAAAAGCAAAGCGAAGCGGGAGCGCAAAAGCCGUUCGAUGCGCAAAAGCAGGGACUCCAAAGAGGAGUC ((((((.((((........))))..((((((......))))))....))))))((((..............)))).........((((((.....)))))) (-28.73 = -30.54 + 1.81)



| Location | 8,231,921 – 8,232,022 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 88.12 |
| Mean single sequence MFE | -38.93 |
| Consensus MFE | -32.97 |
| Energy contribution | -33.35 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8231921 101 - 22224390 GACUCCUCUUUGGAGUCCUUGCUUUUGCGCAUCGAUCGGCUCUGGCGCUCCCGUUUCGCUUUGCUUUUGCGGAAGGAUCCGACCAGGAAUCCGGUGCGGGA ((((((.....)))))).........((((...((.....))..))))((((((.((..(((((....))))).((((((.....)).)))))).)))))) ( -37.20) >DroSec_CAF1 132824 101 - 1 GACUCCUCUUUGGAGUCCCUGCUUUUGCGCAUCGAGCGGCUCUGGCGCUCCCGCUUCGCUUUGUUUUUGCGCAAGGAUCCGACCAGGAACUCGGUGCGGGA ((((((.....))))))((((((((((((((.(((((((....((((....)))))))).)))....)))))))))..((((........)))).))))). ( -41.10) >DroEre_CAF1 121934 101 - 1 GACUCCUCUUUGGAGUCCAUGCACUUGCGCAUCGAACGGCUUUUGCGCUCCCGCUUCGCCUUGCUCUUGCGCAAGGAUCCGACCAGGAUCUCGGUGCGGGA ((((((.....)))))).........(((((............)))))((((((.(((.(((((......)))))(((((.....))))).))).)))))) ( -42.70) >DroYak_CAF1 122994 101 - 1 UUGUCCUCUUUGGAGUCGCUGCUUUUGCGCAUCGAACGACUUUUGCGCUCCCGCCUCGCCUUGCUCUUGCGCAAGGAACCGACCAGGAUCUCGGUGCGGGA ..((((((....)))(((.(((......))).)))..)))........((((((....((((((......)))))).(((((........))))))))))) ( -34.70) >consensus GACUCCUCUUUGGAGUCCCUGCUUUUGCGCAUCGAACGGCUCUGGCGCUCCCGCUUCGCCUUGCUCUUGCGCAAGGAUCCGACCAGGAACUCGGUGCGGGA ((((((.....)))))).........((((...((.....))..))))((((((.(((((((((......)))))).(((.....)))...))).)))))) (-32.97 = -33.35 + 0.38)


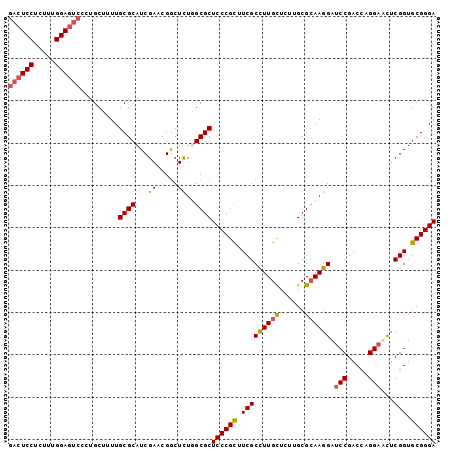
| Location | 8,231,982 – 8,232,085 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.67 |
| Mean single sequence MFE | -39.68 |
| Consensus MFE | -20.98 |
| Energy contribution | -22.46 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8231982 103 + 22224390 GCCGAUCGAUGCGCAAAAGCAAGGACUCCAAAGAGGAGUCUCAUCU---GGUCGAUAUGGGCAUGUUCAGUGGCCGUUCCAUGAGCAUGCCACCCCGUC-----CAUCUGC--------- ((.(((.((((.((....))..(((((((.....))))))))))).---((.((.....((((((((((.(((.....)))))))))))))....)).)-----)))).))--------- ( -39.90) >DroSec_CAF1 132885 103 + 1 GCCGCUCGAUGCGCAAAAGCAGGGACUCCAAAGAGGAGUCCCAUCU---GGUGGACAUGGGCAUGUUCAGUGGCCGUUCCAUGAGCAUGCCAUCCCGUC-----CAUCCGC--------- ((.((.....))))....((.((((((((.....))))))))....---(((((((...((((((((((.(((.....))))))))))))).....)))-----)))).))--------- ( -50.00) >DroEre_CAF1 121995 103 + 1 GCCGUUCGAUGCGCAAGUGCAUGGACUCCAAAGAGGAGUCCCAUCU---GAUGGACAUGGGCAUGUUCAGCGGCCGAUCCAUGAGCAUGCCACCCCGUC-----CAUCCGC--------- .......((((.((....))..(((((((.....))))))))))).---(((((((...((((((((((..((.....)).)))))))))).....)))-----))))...--------- ( -45.70) >DroWil_CAF1 131861 109 + 1 -UUAAAUGAUUUGGAAAUAC-------CGCAAGAUGAUAAUGAUAA---UGGCGAUAUGACAUUGUUUAGUGGUCGCUCAAUGAGUUUGCCACCAAGGCCCCUGCAUUCGGCGCUAAAAC -........(((((....((-------(((.....(((((((...(---((....)))..)))))))..)))))((((.((((.....(((.....))).....)))).))))))))).. ( -23.00) >DroYak_CAF1 123055 106 + 1 GUCGUUCGAUGCGCAAAAGCAGCGACUCCAAAGAGGACAACCACCUGCUGGUGGACAUGGGCAUGUUCAGUGGCCGCUCCAUGAGCAUGCCACCCCGAC-----CAUCUGC--------- (((((....(((......)))))))).....(((((....(((((....))))).....((((((((((.(((.....))))))))))))).......)-----).)))..--------- ( -39.80) >consensus GCCGAUCGAUGCGCAAAAGCAGGGACUCCAAAGAGGAGUCCCAUCU___GGUGGACAUGGGCAUGUUCAGUGGCCGCUCCAUGAGCAUGCCACCCCGUC_____CAUCCGC_________ .......((((............((((((.....))))))(((((....))))).....((((((((((.(((.....))))))))))))).............))))............ (-20.98 = -22.46 + 1.48)

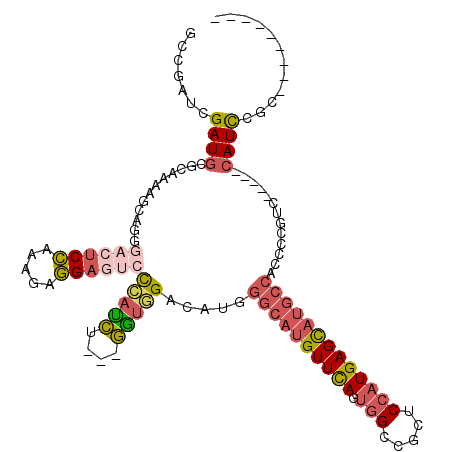
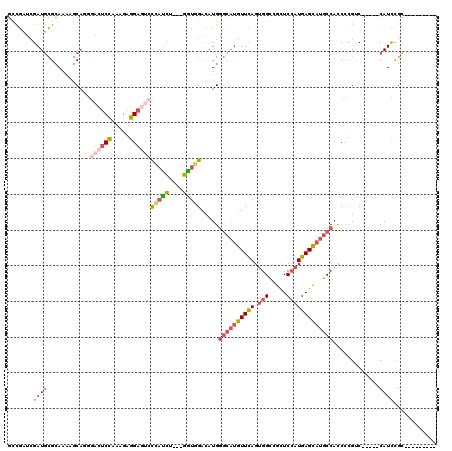
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:33 2006