
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,223,267 – 8,223,369 |
| Length | 102 |
| Max. P | 0.996682 |

| Location | 8,223,267 – 8,223,369 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -28.38 |
| Energy contribution | -29.13 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
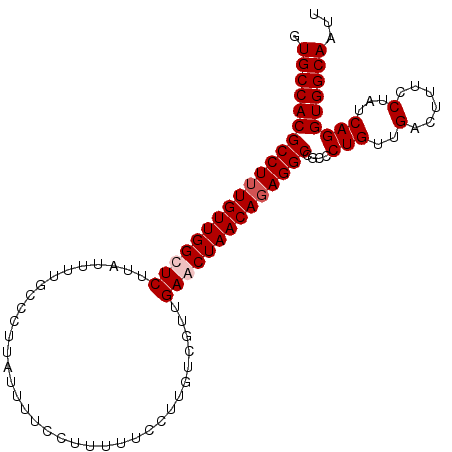
>X_DroMel_CAF1 8223267 102 + 22224390 GUGCCACGCCUUUGUUGGCUCUUAUUUUGGCCUUAUUUUCCUUUUUCCUUGUCGUUGAACUAACAGAGGCCGCCCUGUUGACUUUCCUAUCAGGUGGCAAUU .(((((((((((((((((.((......((((...................))))..)).)))))))))))....(((..(......)...)))))))))... ( -28.51) >DroSec_CAF1 124396 102 + 1 GUGCCACGCCUUUGUUGGUUCUUAUUUUGGCCUUAUUUUCCUUUUUCCCUGUCGUUGAACUAACAGAGGCCGCCCUGUUGACUUUCCUAUCAGGUGGCAAUU .((((((((((((((((((((.......((.................)).......))))))))))))))....(((..(......)...)))))))))... ( -32.77) >DroEre_CAF1 113803 102 + 1 GUGCCACGCCUUUGUUGGUUCCUAUUUUGCCAGUAUUUUCCUUUUUCCUUGUCGUUGAACUAACAGAGGCCUUCCUGUUGACUUUCCUAUCAGGUGGCAAUU .((((((((((((((((((((...................................))))))))))))))....(((..(......)...)))))))))... ( -30.25) >DroYak_CAF1 114792 102 + 1 GUGCCACGCCUUUGUUGGCUCUUAUUUGGCCCCUAUUUUCCUUUUUCCUUGUCGUUGAACUAACACAGGCCGCCCUGUUGACUUUCCUAUCAGGUGGCAAUU .((((((((((.((((((.((.....((((....................))))..)).)))))).))))....(((..(......)...)))))))))... ( -25.45) >consensus GUGCCACGCCUUUGUUGGCUCUUAUUUUGCCCUUAUUUUCCUUUUUCCUUGUCGUUGAACUAACAGAGGCCGCCCUGUUGACUUUCCUAUCAGGUGGCAAUU .((((((((((((((((((((...................................))))))))))))))....(((..(......)...)))))))))... (-28.38 = -29.13 + 0.75)

| Location | 8,223,267 – 8,223,369 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -26.19 |
| Consensus MFE | -23.48 |
| Energy contribution | -23.23 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8223267 102 - 22224390 AAUUGCCACCUGAUAGGAAAGUCAACAGGGCGGCCUCUGUUAGUUCAACGACAAGGAAAAAGGAAAAUAAGGCCAAAAUAAGAGCCAACAAAGGCGUGGCAC ...((((((...........(((.....))).((((.((((.((((..(.....)......((.........)).......)))).)))).)))))))))). ( -25.70) >DroSec_CAF1 124396 102 - 1 AAUUGCCACCUGAUAGGAAAGUCAACAGGGCGGCCUCUGUUAGUUCAACGACAGGGAAAAAGGAAAAUAAGGCCAAAAUAAGAACCAACAAAGGCGUGGCAC ...((((((...........(((.....))).((((.((((.((((..(....).......((.........)).......)))).)))).)))))))))). ( -25.70) >DroEre_CAF1 113803 102 - 1 AAUUGCCACCUGAUAGGAAAGUCAACAGGAAGGCCUCUGUUAGUUCAACGACAAGGAAAAAGGAAAAUACUGGCAAAAUAGGAACCAACAAAGGCGUGGCAC ...((((((.((((......))))........((((.((((.((((..(.....)..............(((......))))))).)))).)))))))))). ( -25.50) >DroYak_CAF1 114792 102 - 1 AAUUGCCACCUGAUAGGAAAGUCAACAGGGCGGCCUGUGUUAGUUCAACGACAAGGAAAAAGGAAAAUAGGGGCCAAAUAAGAGCCAACAAAGGCGUGGCAC ...((((((...........(((.....)))(((((.((((..(((...............))).)))).)))))........(((......))))))))). ( -27.86) >consensus AAUUGCCACCUGAUAGGAAAGUCAACAGGGCGGCCUCUGUUAGUUCAACGACAAGGAAAAAGGAAAAUAAGGCCAAAAUAAGAACCAACAAAGGCGUGGCAC ...((((((.((((......))))........((((.((((.((((...................................)))).)))).)))))))))). (-23.48 = -23.23 + -0.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:27 2006