
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,221,830 – 8,221,928 |
| Length | 98 |
| Max. P | 0.715213 |

| Location | 8,221,830 – 8,221,928 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -22.85 |
| Consensus MFE | -17.42 |
| Energy contribution | -17.87 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
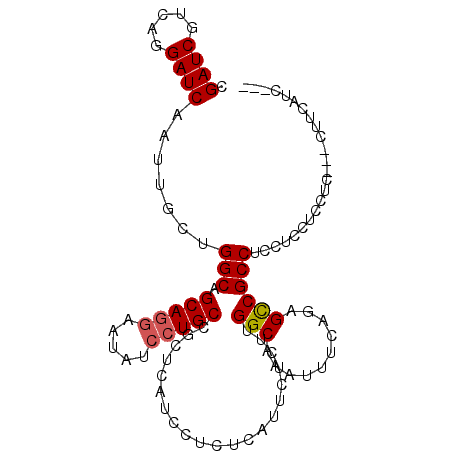
>X_DroMel_CAF1 8221830 98 + 22224390 CGAUCGUAAGGAUCAAUUGCCGGCAGCAGGAAUAUCCUGCCGCUCAUCCUCUCAUUCUCUUGGCAAAUUUCAGAGCCGCCUCCUGCUCCUC---CUUCAUC--- .(((.(.(((((....((((((((.(((((.....))))).))).................)))))......((((........)))).))---)))))))--- ( -25.92) >DroSec_CAF1 123077 98 + 1 CGAUCGUCAGGAUCAAUUGCUGGCAGCAGGAAUAUCCUGCCGCUCAUCCUCUCAUUCUGUUGGCAAAUUUCAGAGCCGCCGCCUCCUGCUC---CUCCAUC--- .((((.....))))..((((..((((((((.....))))).................)))..))))......((((.(.......).))))---.......--- ( -23.72) >DroYak_CAF1 113287 102 + 1 CGAUCGUCAGGAUCAAUUGCUGGCAGCAAAAAUAUCCUGCCGCUCAUCU--GCAAUCUCUUGGCAAAUUUCAGAGUCGCCUCCACUUCCACUUCCAUUUCCACC .....(((((((...(((((((((((..........)))))).......--))))).)))))))........(((....)))...................... ( -18.91) >consensus CGAUCGUCAGGAUCAAUUGCUGGCAGCAGGAAUAUCCUGCCGCUCAUCCUCUCAUUCUCUUGGCAAAUUUCAGAGCCGCCUCCUCCUCCUC___CUUCAUC___ .((((.....)))).......(((.(((((.....))))).....................(((..........))))))........................ (-17.42 = -17.87 + 0.45)

| Location | 8,221,830 – 8,221,928 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -22.68 |
| Energy contribution | -21.80 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
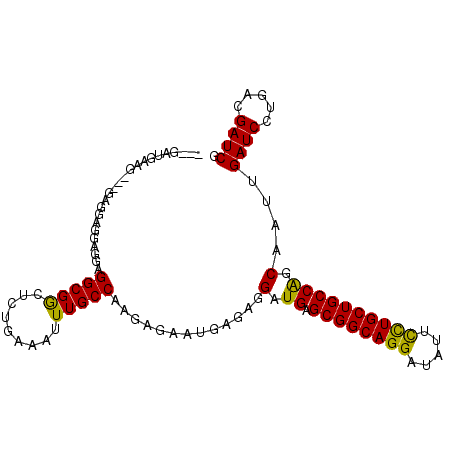
>X_DroMel_CAF1 8221830 98 - 22224390 ---GAUGAAG---GAGGAGCAGGAGGCGGCUCUGAAAUUUGCCAAGAGAAUGAGAGGAUGAGCGGCAGGAUAUUCCUGCUGCCGGCAAUUGAUCCUUACGAUCG ---(((.(((---(((((((........))))).....(((((.......(....)...(.((((((((.....))))))))))))))....)))))...))). ( -34.40) >DroSec_CAF1 123077 98 - 1 ---GAUGGAG---GAGCAGGAGGCGGCGGCUCUGAAAUUUGCCAACAGAAUGAGAGGAUGAGCGGCAGGAUAUUCCUGCUGCCAGCAAUUGAUCCUGACGAUCG ---(((.(((---((.(((...(((((((.........)))))...............((.((((((((.....))))))))))))..))).))))..).))). ( -33.10) >DroYak_CAF1 113287 102 - 1 GGUGGAAAUGGAAGUGGAAGUGGAGGCGACUCUGAAAUUUGCCAAGAGAUUGC--AGAUGAGCGGCAGGAUAUUUUUGCUGCCAGCAAUUGAUCCUGACGAUCG (((.(....(((.(..((..(((((....)))))...))..).....((((((--...((.(((((((((...)))))))))))))))))..)))...).))). ( -29.30) >consensus ___GAUGAAG___GAGGAGGAGGAGGCGGCUCUGAAAUUUGCCAAGAGAAUGAGAGGAUGAGCGGCAGGAUAUUCCUGCUGCCAGCAAUUGAUCCUGACGAUCG ........................(((((.........))))).............(.((.((((((((.....)))))))))).)....((((.....)))). (-22.68 = -21.80 + -0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:25 2006