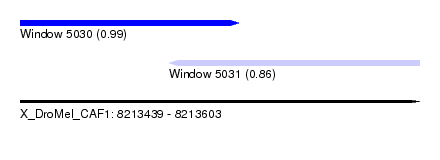
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,213,439 – 8,213,603 |
| Length | 164 |
| Max. P | 0.992325 |

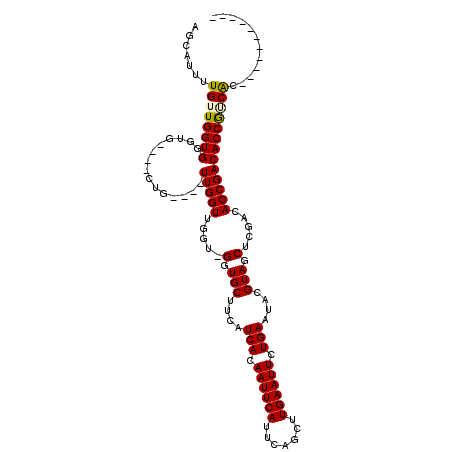
| Location | 8,213,439 – 8,213,529 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 84.01 |
| Mean single sequence MFE | -39.10 |
| Consensus MFE | -27.32 |
| Energy contribution | -29.31 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8213439 90 + 22224390 GGAUGCGGACACAUCCACGGAUUUGGAUCCGGAUUCGGAUACGGAUUCAGAUUCGGUGACG------------UCGACGGUGUCGGUGUCGAGCUACGUAUU (((((......))))).((((((((((((((..........)))))))))))))).((((.------------.((((...))))..))))........... ( -36.50) >DroSec_CAF1 114824 90 + 1 GGAUGCGGACACAUCCACGGAUUUGGAUCCGGAUUCGGAUGCGGAUUCAGAUUCGGUGACG------------GUGACGGUGUCGGUGUCGAGCUACGUAUU .(((((.(((((.((..((((((((((((((.((....)).))))))))))))))..))((------------....))))))).)))))............ ( -40.70) >DroEre_CAF1 104894 102 + 1 GGAUGCGGACACAUCCACGGAUUUGGAUCCGGAUUCGGAUUCGGAUUCAGAUUCGUUGACGGUGUCGUUGUCGGUGUCGGUGUCGGUGUCGAGCUACGUAUU .(((((.(((((.((.((((((((((((((((((....)))))))))))))))))).))(((..(((....)))..)))))))).)))))............ ( -47.60) >DroYak_CAF1 105482 90 + 1 GGAUGCGGACACAUCCACGGAUUUGGAUACGUAUUCGGAU------UCCGUUUCGGUGACGGUG------AUGGUGCUGGUGUCGGUGUCGAGCUACGUAUU .(((((.(((((...(((((((((((((....))))))))------))(((((((....))).)------))))))...))))).)))))............ ( -31.60) >consensus GGAUGCGGACACAUCCACGGAUUUGGAUCCGGAUUCGGAU_CGGAUUCAGAUUCGGUGACG____________GUGACGGUGUCGGUGUCGAGCUACGUAUU .(((((.(((((...((((((((((((((((.((....)).)))))))))))))).)).(((..............)))))))).)))))............ (-27.32 = -29.31 + 2.00)

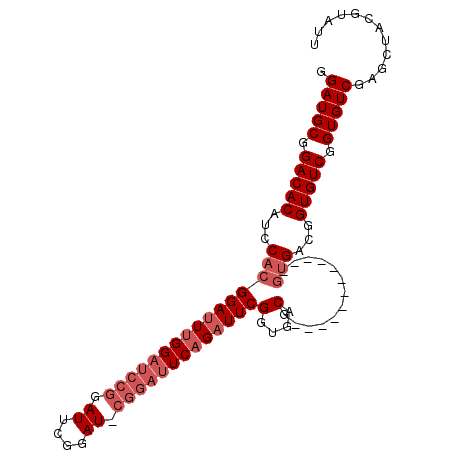
| Location | 8,213,500 – 8,213,603 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.34 |
| Mean single sequence MFE | -33.15 |
| Consensus MFE | -19.20 |
| Energy contribution | -19.26 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8213500 103 - 22224390 AGCAUUUUGUUGGUGG-UGCAACCAACAGCUUGGUGGGUGGGUGCUUCAUCACAAUUCAUUCAGCUUGAAUUCUGAAUACGUAGCUCGACACCGACACCGUCGA----------- ......((((((((..-....)))))))).(((..((((.((((.....(((.((((((.......)))))).)))...((.....)).)))).)).))..)))----------- ( -30.10) >DroSec_CAF1 114885 96 - 1 AGCAUUUUGUUGGUGGGUGU---UGGUUGCUUGGU-----GGUGCUUCAUCACAAUUCAUUCAGCUUGAAUUCUGAAUACGUAGCUCGACACCGACACCGUCAC----------- .......((.((((((((((---(((((((...((-----((((...)))))).....((((((........))))))..))))).)))))))..))))).)).----------- ( -32.20) >DroEre_CAF1 104956 103 - 1 AGCAUUUUGUUGGUGGG-------UG----UUGGUUGGU-GGUGCUUCAUCACAAUUCAUUCAGCUUGAAUUCUGAAUACGUAGCUCGACACCGACACCGACACCGACAACGACA ......(((((((((((-------((----(((((..((-.(.(((...(((.((((((.......)))))).)))......))).).)))))))))))..)))))))))..... ( -37.40) >DroYak_CAF1 105538 101 - 1 AGCAUUUUGUUGGUGGGUUG---CUG----UUGGUUGGU-GGUGCUUCAUCACAAUUCAUUCAGCUUGAAUUCUGAAUACGUAGCUCGACACCGACACCAGCACCAU------CA ..........((((((..((---(((----...((((((-(..(((...(((.((((((.......)))))).)))......)))....)))))))..)))))))))------)) ( -32.90) >consensus AGCAUUUUGUUGGUGGGUG____CUG____UUGGUUGGU_GGUGCUUCAUCACAAUUCAUUCAGCUUGAAUUCUGAAUACGUAGCUCGACACCGACACCGUCAC___________ .......((((((((...............(((((.....(.(((....(((.((((((.......)))))).)))....))).).....)))))))))))))............ (-19.20 = -19.26 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:23 2006