| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,213,138 – 8,213,230 |
| Length | 92 |
| Max. P | 0.982443 |

| Location | 8,213,138 – 8,213,230 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 83.47 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -27.49 |
| Energy contribution | -28.05 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
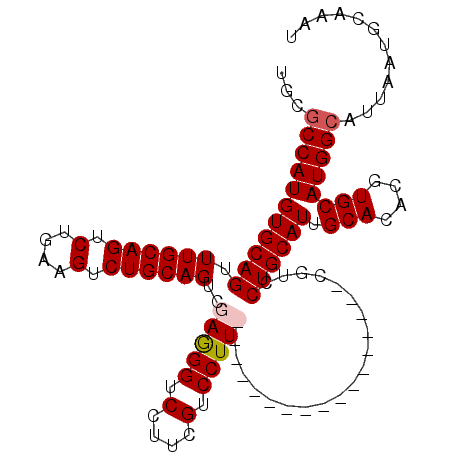
>X_DroMel_CAF1 8213138 92 + 22224390 UGCGCCAUGUGCAGUUUGCAGUCUGAAGUCUGCAGUCGAGGGUCCUUCGUCCUU-----------------CGUCCUGCAUUGCACACGUGCAUGGCAUUAAUGCAAAU ...((((((..(.((.(((((.........(((((.((((((.(....).))))-----------------))..)))))))))).)))..))))))............ ( -35.20) >DroSec_CAF1 114578 79 + 1 UGCGCCAUGUGCAGUUUGCAGUCUGAAGUCUGCAGUCGAGGGUCCUUCGUCCUU-----------------CGUCCUGCAUUGCACACGUGCAUGG------------- (((((..((((((((.(((((.(....).)))))(.((((((.(....).))))-----------------)).)....)))))))).)))))...------------- ( -31.20) >DroEre_CAF1 104628 99 + 1 UGCGCCAUGUGCAGUUUGCAGUCUGAAGUCUGCAGUCGAGGGUCCUUCGUCCUUCAUCCUU----------CGUCCUGCAUUGCACACGUGCAUGGCAUUAAUGCAAAU (((((..(((((((..(((((..(((((..((.((.(((((...)))))..)).))..)))----------))..)))))))))))).)))))..(((....))).... ( -35.60) >DroYak_CAF1 105177 109 + 1 UGCGCCAUGUGCAGUUUGCAGUCUGAAGUCUGCAGUCGAAGGUCCUUCGUCCUUCGUCGUUCGCCUCUCGUCGUCCUGCAUUGCACACGUGCAUGGCAUUAAUGCAAAU (((((..(((((((..(((((..(((.....((.(.((((((.(....).)))))).)....))......)))..)))))))))))).)))))..(((....))).... ( -36.30) >consensus UGCGCCAUGUGCAGUUUGCAGUCUGAAGUCUGCAGUCGAGGGUCCUUCGUCCUU_________________CGUCCUGCAUUGCACACGUGCAUGGCAUUAAUGCAAAU ...(((((((((((.((((((.(....).))))))..(((((.(....).)))))....................)))))).(((....))))))))............ (-27.49 = -28.05 + 0.56)


| Location | 8,213,138 – 8,213,230 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 83.47 |
| Mean single sequence MFE | -32.17 |
| Consensus MFE | -24.70 |
| Energy contribution | -24.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8213138 92 - 22224390 AUUUGCAUUAAUGCCAUGCACGUGUGCAAUGCAGGACG-----------------AAGGACGAAGGACCCUCGACUGCAGACUUCAGACUGCAAACUGCACAUGGCGCA ....(((....)))..(((.((((((((.(((((..((-----------------(.((.(....).)).))).(((.......))).)))))...))))))))..))) ( -29.90) >DroSec_CAF1 114578 79 - 1 -------------CCAUGCACGUGUGCAAUGCAGGACG-----------------AAGGACGAAGGACCCUCGACUGCAGACUUCAGACUGCAAACUGCACAUGGCGCA -------------...(((.((((((((.(((((..((-----------------(.((.(....).)).))).(((.......))).)))))...))))))))..))) ( -28.90) >DroEre_CAF1 104628 99 - 1 AUUUGCAUUAAUGCCAUGCACGUGUGCAAUGCAGGACG----------AAGGAUGAAGGACGAAGGACCCUCGACUGCAGACUUCAGACUGCAAACUGCACAUGGCGCA ....(((....)))..(((.((((((((.(((((...(----------(((..((.((..(((.(....)))).)).))..))))...)))))...))))))))..))) ( -34.10) >DroYak_CAF1 105177 109 - 1 AUUUGCAUUAAUGCCAUGCACGUGUGCAAUGCAGGACGACGAGAGGCGAACGACGAAGGACGAAGGACCUUCGACUGCAGACUUCAGACUGCAAACUGCACAUGGCGCA ....(((....)))..(((.((((((((.(((((..((.(....).)).....((((((.(....).))))))...............)))))...))))))))..))) ( -35.80) >consensus AUUUGCAUUAAUGCCAUGCACGUGUGCAAUGCAGGACG_________________AAGGACGAAGGACCCUCGACUGCAGACUUCAGACUGCAAACUGCACAUGGCGCA ................(((.((((((((.(((((.......................((.(....).)).....(((.......))).)))))...))))))))..))) (-24.70 = -24.70 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:21 2006