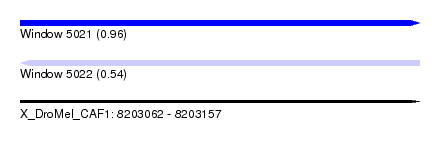
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,203,062 – 8,203,157 |
| Length | 95 |
| Max. P | 0.961543 |

| Location | 8,203,062 – 8,203,157 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 76.46 |
| Mean single sequence MFE | -29.16 |
| Consensus MFE | -11.88 |
| Energy contribution | -13.92 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8203062 95 + 22224390 UCGCCU--C-AUCCUUGCGGAGGCCAUUCAGCAUUUCAGCUUCGAUUAAUGUUGCCAAAAAAAAA----CCGGGCAUCUGGGAAGAGCUGAGAUGUCUGGCU------ ..((((--(-.........)))))...(((((((((((((((((((....))))...........----((((....))))...))))))))))).))))..------ ( -30.60) >DroSec_CAF1 105054 87 + 1 UCGCUCCUCAAUCCUUGCGGCGGCCAUUCAGCAUUUCAGCUUCGAUUAAUGUUGCCAAAAAA-------UCG--------------GCUGAGAUGUCUGGCCGUACAA .(((............)))(((((((....((((((((((..(((((..((....))...))-------)))--------------)))))))))).))))))).... ( -30.80) >DroSim_CAF1 84899 87 + 1 ACGCUCCUCAAUCCUUGCGGCGGCCAUUCAGCAUUUCAGCUUCGAUUAAUGUUGCCAAAAAA-------UCG--------------GCUGAGAUGUCUGGCCGUACAA .(((............)))(((((((....((((((((((..(((((..((....))...))-------)))--------------)))))))))).))))))).... ( -31.20) >DroEre_CAF1 95312 97 + 1 UCACUCCGC-UUCCUUGCGGAGG-CAUUCAGCAUUUCAGCUUCGAUUAAUGUUGCCAAAAA--------UCGGACAGAUAGGAA-AACAGAGAUGUCUGGGCGUAUAA ...((((((-......))))))(-(.(((((((((((.(.(((((((..((....))..))--------)))))).....(...-..).)))))).))))).)).... ( -27.90) >DroYak_CAF1 95163 105 + 1 UCACUCAUC-AUCCUUGCGGAGG-CAUUCAGCAUUUCGGCUUCGAUUAAUGUUGCCAAAAAAAAAAAAUUGGGGCAGAUAGGAA-AGCAGAGAUGUCUGGGCGUACAA ...(((..(-(....))..)))(-(.(((((((((((.((((...(((...(((((..((........))..))))).)))..)-))).)))))).))))).)).... ( -25.30) >consensus UCGCUCCUC_AUCCUUGCGGAGGCCAUUCAGCAUUUCAGCUUCGAUUAAUGUUGCCAAAAAA_______UCGG_CA__U_GGAA_AGCUGAGAUGUCUGGCCGUACAA ...........(((....)))(((((....((((((((((..((((....))))((...............)).............)))))))))).)))))...... (-11.88 = -13.92 + 2.04)


| Location | 8,203,062 – 8,203,157 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 76.46 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -13.69 |
| Energy contribution | -14.33 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8203062 95 - 22224390 ------AGCCAGACAUCUCAGCUCUUCCCAGAUGCCCGG----UUUUUUUUUGGCAACAUUAAUCGAAGCUGAAAUGCUGAAUGGCCUCCGCAAGGAU-G--AGGCGA ------...(((.(((.((((((((....)))((((..(----......)..))))............))))).))))))....((((((....))).-.--.))).. ( -27.50) >DroSec_CAF1 105054 87 - 1 UUGUACGGCCAGACAUCUCAGC--------------CGA-------UUUUUUGGCAACAUUAAUCGAAGCUGAAAUGCUGAAUGGCCGCCGCAAGGAUUGAGGAGCGA ((((.((((((..(((.(((((--------------(((-------((...((....))..)))))..))))).))).....))))))((....))........)))) ( -31.50) >DroSim_CAF1 84899 87 - 1 UUGUACGGCCAGACAUCUCAGC--------------CGA-------UUUUUUGGCAACAUUAAUCGAAGCUGAAAUGCUGAAUGGCCGCCGCAAGGAUUGAGGAGCGU ..((.((((((..(((.(((((--------------(((-------((...((....))..)))))..))))).))).....))))))((....))........)).. ( -31.00) >DroEre_CAF1 95312 97 - 1 UUAUACGCCCAGACAUCUCUGUU-UUCCUAUCUGUCCGA--------UUUUUGGCAACAUUAAUCGAAGCUGAAAUGCUGAAUG-CCUCCGCAAGGAA-GCGGAGUGA ...(((.(((((.(((.((.(((-(...........(((--------((..((....))..))))))))).)).))))))...(-(.(((....))).-)))).))). ( -27.60) >DroYak_CAF1 95163 105 - 1 UUGUACGCCCAGACAUCUCUGCU-UUCCUAUCUGCCCCAAUUUUUUUUUUUUGGCAACAUUAAUCGAAGCCGAAAUGCUGAAUG-CCUCCGCAAGGAU-GAUGAGUGA .....(((.((..((((..(((.-........................(((((((..(.......)..))))))).((.....)-)....)))..)))-).)).))). ( -21.50) >consensus UUGUACGGCCAGACAUCUCAGCU_UUCC_A__UG_CCGA_______UUUUUUGGCAACAUUAAUCGAAGCUGAAAUGCUGAAUGGCCUCCGCAAGGAU_GAGGAGCGA .........(((.(((.((((((........................((..((....))..))....)))))).))))))........((....))............ (-13.69 = -14.33 + 0.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:15 2006