| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,201,217 – 8,201,323 |
| Length | 106 |
| Max. P | 0.844579 |

| Location | 8,201,217 – 8,201,323 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.53 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -7.74 |
| Energy contribution | -8.46 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.27 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.23 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8201217 106 - 22224390 UUUGUUUUUGAACGCACCAUUUGCUUGGAGGGGGCGGGGUGGUUUU----UUCGCCCAGCCA-AAUGUUG---UGUUGGAAA-----AAGCCAGGGAUAUUAGUUUUAUUCUG-GGAUCA ....(((((.((((((.((((....(((...(((((..(......)----..)))))..)))-)))).))---)))).))))-----)..((.((((((.......)))))).-)).... ( -35.30) >DroSec_CAF1 103289 101 - 1 UUUGUUUUUGAACGCACCAUUUGCUUGGGGG-----GGUUGCUUUU----UUCGCCCAGCCA-AAUGUUG---UGUUGGAAA-----AUGCCUGGGAUAUUAGUUUUAUUCCG-GGAUCA ...((((((.((((((.((((((.(((((.(-----((........----))).))))).))-)))).))---)))).))))-----)).(((((((((.......)))))))-)).... ( -40.50) >DroSim_CAF1 83062 101 - 1 UUUGUUUUUGAACGCCCCAUUUGCUUGGGGG-----GUUUGGUUUU----UUUGCCCAGCCA-AAUGUUG---UGUUGGAAA-----AUGCCUGGGAUAUUAGUUUUAUUCUG-GGAUCA ...((((((.(((.(((((......)))))(-----(((.(((...----...))).)))).-.......---.))).))))-----)).((..(((((.......)))))..-)).... ( -33.00) >DroEre_CAF1 93497 95 - 1 UUUGUUUUUGAACGCCCCAUUUGCUCGGGC-----------UUCCU----UUCGCCCAUCCA-AAUGUUG---UGUUGGAAA-----AAGCCUGGGAUAUUAGUUUUAUUCUG-GGAUCA ....(((((.(((((..((((((...((((-----------.....----...))))...))-))))..)---)))).))))-----)..((..(((((.......)))))..-)).... ( -30.30) >DroYak_CAF1 92774 96 - 1 UUUGUUUUUGAACGCCCCAUUUGCUCGGGG-----------GUCCU----UUGGCCCAUCCAAAAUGUUG---UGUUGGAAA-----AAGCCUGGGAUAUUAGUUUCAUUCCG-GGAUCA ....(((((.(((((..(((((......((-----------(((..----..))))).....)))))..)---)))).))))-----)..((((((((.........))))))-)).... ( -28.60) >DroAna_CAF1 142898 108 - 1 UUU---GUUGCAUGCCCCAUUUGGCACAGG--------GUCCUUUUCAUUCCUUCCCCCUCA-GGCGUUAUCCUGUUGGGAUUCCUGCACCCUGGGAUAUUAGUUUUAUUCCGGGGAUCA ...---..((((((((......)))).(((--------(..........))))..(((..((-((......))))..))).....))))((((((((((.......)))))))))).... ( -34.70) >consensus UUUGUUUUUGAACGCCCCAUUUGCUUGGGG_________UGGUUUU____UUCGCCCAGCCA_AAUGUUG___UGUUGGAAA_____AAGCCUGGGAUAUUAGUUUUAUUCCG_GGAUCA ......(((.((((...((((.(((.(((.........................))))))...))))......)))).))).........(((((((((.......))))))).)).... ( -7.74 = -8.46 + 0.72)


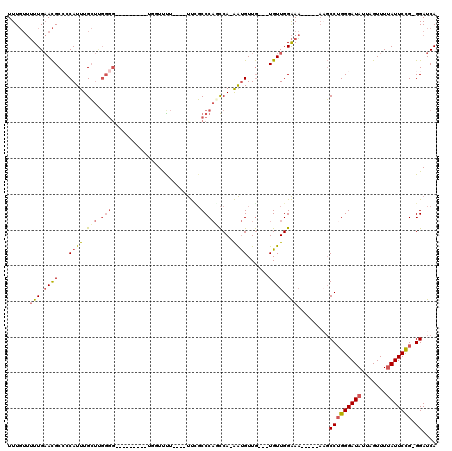
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:13 2006