
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,196,271 – 8,196,386 |
| Length | 115 |
| Max. P | 0.611970 |

| Location | 8,196,271 – 8,196,386 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.75 |
| Mean single sequence MFE | -32.95 |
| Consensus MFE | -14.01 |
| Energy contribution | -16.15 |
| Covariance contribution | 2.14 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.43 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
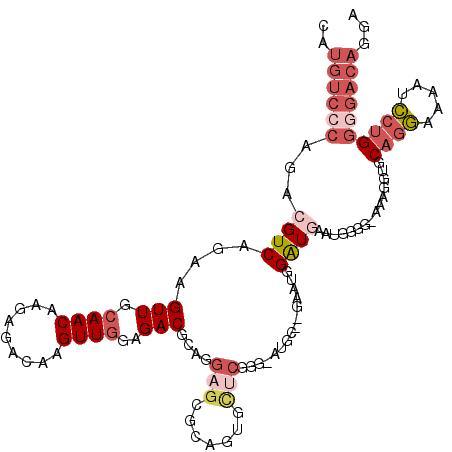
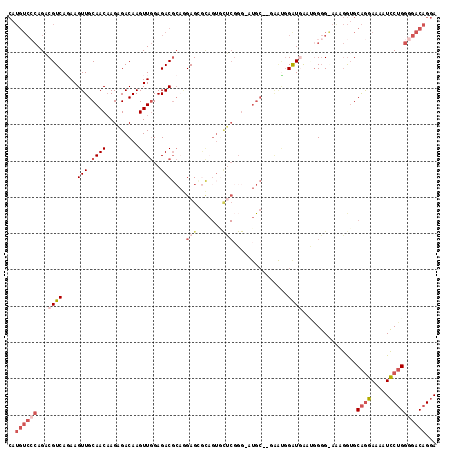
>X_DroMel_CAF1 8196271 115 + 22224390 CAUGUCCCAGACGUCAGAAGUUGCAACAAGAGACAAGUUGGAGACGCAGGAGCGCAGUGCUCGGG-AUGC--GAAUGGAUGAAUAGGGGAAAGGUGCAGGAAAAUCCUGGGGACAGGA ..((((((...((((.....(((((.(..(((((.....(....)((......)).)).)))..)-.)))--))...))))...............((((.....))))))))))... ( -34.90) >DroSec_CAF1 98517 114 + 1 CAUGUCCCAGACGUCAGAAGUUGCAACCAGAGACAAGUUGGAGACGCAGGAGCACAGUGCUCGGG-AUGC--GAAUGGAUGAAUGGGG-AGAGGUGCAGGAAAAUCCUGGGGACAGGA ....(((((..((((.....(((((.((.(((((.....(....)((....))...)).))).))-.)))--))...))))..)))))-...............(((((....))))) ( -38.10) >DroSim_CAF1 78287 115 + 1 CAUGUCCCAGACGUCAGAAGUUGCAACAAGAGACAAGUUGGAGACGCAGGAGCGCAGUGCUCGGG-AUGC--GAAUGGAUGAAUGGGGAAGAGGUGCAGGAAAAUCCUGGGGACAGGA ....(((((..((((.....(((((.(..(((((.....(....)((......)).)).)))..)-.)))--))...))))..)))))................(((((....))))) ( -36.10) >DroEre_CAF1 88653 116 + 1 CAUGUCCCAGACGUCACAAGUUGCAACAAGAGACAAGUUGGAGACGCUAGAGAGCAGUACUCGGGGCUGCAAGAAUGGAUGG--GGGGGAAAGGUGCAGGAAAAUCCUGGGGACAGGA (((.((((...((((.((..(((((.(..(((((.....(....)(((....))).)).)))..)..)))))...)))))).--..))))...)))........(((((....))))) ( -37.10) >DroYak_CAF1 87601 104 + 1 CAUGUCGCAGCCGUCAGAAGUUGCAACAAGAGACAAGUUGGAGACGCAAGAGAGU------------UGCAAAGGCUGGUUG--GGGGAAAAGGUGCAGGAAAAUCCUGGGGACAGGA ..(.((.((((((((.....(((((((....((....))(....)........))------------))))).))).)))))--.)).)...............(((((....))))) ( -29.30) >DroAna_CAF1 136764 95 + 1 --------AGCCGUCACAAGUUGCAACAAGCGACAAGUUAGAGACCA--GAGG---GUGUGCGGG-G--------GGGAUUGGCGGCG-AGACAAGCGACAGGAUGCAGUGAACAGGA --------.(((((((...(((((.....))))).......(.(((.--...)---)).).....-.--------.....))))))).-......(((......)))........... ( -22.20) >consensus CAUGUCCCAGACGUCAGAAGUUGCAACAAGAGACAAGUUGGAGACGCAGGAGCGCAGUGCUCGGG_AUGC__GAAUGGAUGAAUGGGG_AAAGGUGCAGGAAAAUCCUGGGGACAGGA ..((((((...((((....(((.((((.........))))..)))....(((.......)))...............))))...............((((.....))))))))))... (-14.01 = -16.15 + 2.14)

| Location | 8,196,271 – 8,196,386 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.75 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -13.74 |
| Energy contribution | -14.77 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
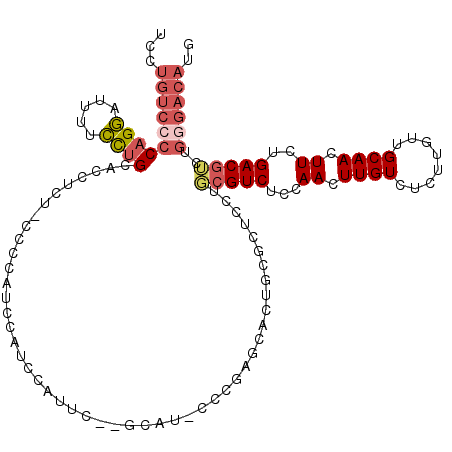
>X_DroMel_CAF1 8196271 115 - 22224390 UCCUGUCCCCAGGAUUUUCCUGCACCUUUCCCCUAUUCAUCCAUUC--GCAU-CCCGAGCACUGCGCUCCUGCGUCUCCAACUUGUCUCUUGUUGCAACUUCUGACGUCUGGGACAUG ...((((((((((.....))))........................--....-...((((.....))))..(((((...((.((((........)))).))..)))))..)))))).. ( -25.00) >DroSec_CAF1 98517 114 - 1 UCCUGUCCCCAGGAUUUUCCUGCACCUCU-CCCCAUUCAUCCAUUC--GCAU-CCCGAGCACUGUGCUCCUGCGUCUCCAACUUGUCUCUGGUUGCAACUUCUGACGUCUGGGACAUG ...((((((((((.....)))).......-...............(--(((.-...((((.....)))).))))..........(((...(((....)))...)))....)))))).. ( -26.70) >DroSim_CAF1 78287 115 - 1 UCCUGUCCCCAGGAUUUUCCUGCACCUCUUCCCCAUUCAUCCAUUC--GCAU-CCCGAGCACUGCGCUCCUGCGUCUCCAACUUGUCUCUUGUUGCAACUUCUGACGUCUGGGACAUG ...((((((((((.....))))........................--....-...((((.....))))..(((((...((.((((........)))).))..)))))..)))))).. ( -25.00) >DroEre_CAF1 88653 116 - 1 UCCUGUCCCCAGGAUUUUCCUGCACCUUUCCCCC--CCAUCCAUUCUUGCAGCCCCGAGUACUGCUCUCUAGCGUCUCCAACUUGUCUCUUGUUGCAACUUGUGACGUCUGGGACAUG ...((((((((((.....))))............--.....(((..(((((((...(((.((.(((....)))((.....))..)))))..)))))))...)))......)))))).. ( -27.40) >DroYak_CAF1 87601 104 - 1 UCCUGUCCCCAGGAUUUUCCUGCACCUUUUCCCC--CAACCAGCCUUUGCA------------ACUCUCUUGCGUCUCCAACUUGUCUCUUGUUGCAACUUCUGACGGCUGCGACAUG ...((((..((((.....))))............--....(((((...(((------------(.....))))(((...((.((((........)))).))..)))))))).)))).. ( -22.70) >DroAna_CAF1 136764 95 - 1 UCCUGUUCACUGCAUCCUGUCGCUUGUCU-CGCCGCCAAUCCC--------C-CCCGCACAC---CCUC--UGGUCUCUAACUUGUCGCUUGUUGCAACUUGUGACGGCU-------- ...........((........))......-.(((((((.....--------.-.........---....--)))).........(((((..((....))..)))))))).-------- ( -14.71) >consensus UCCUGUCCCCAGGAUUUUCCUGCACCUCU_CCCCAUCCAUCCAUUC__GCAU_CCCGAGCACUGCGCUCCUGCGUCUCCAACUUGUCUCUUGUUGCAACUUCUGACGUCUGGGACAUG ...((((((((((.....)))).................................................(((((...((.((((........)))).))..)))))..)))))).. (-13.74 = -14.77 + 1.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:11 2006