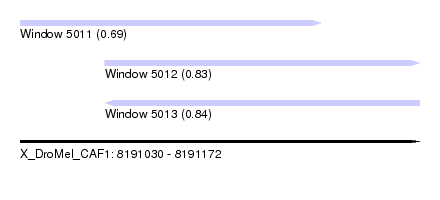
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,191,030 – 8,191,172 |
| Length | 142 |
| Max. P | 0.837764 |

| Location | 8,191,030 – 8,191,137 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.97 |
| Mean single sequence MFE | -41.16 |
| Consensus MFE | -30.72 |
| Energy contribution | -31.48 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8191030 107 + 22224390 ---CCCCUUC--CGUUAUCCGGUAGCCA-----UUUGCAUGCCGUCCACGAGGCG-GUGGCACAGAUUUCUGGACUCGAAGUCUGAGCCUGCGACUGAGCCACCACUAGAAUCUGGCC-- ---.......--........(((.((((-----.(((((.((((((.....))))-))(((.((((((((.......)))))))).)))))))).)).)).))).((((...))))..-- ( -36.80) >DroSec_CAF1 93496 110 + 1 CCCCCCCUUC--CGCCAUCCGGUGGCCA-----UUUGCAUGCCGUCCACGAGGCG-GUGGCACAGAUUUCUGGACUCGAAGUCUGAGCCUGCGACUGAGCCACCACUAGAAUCUGGCC-- ..........--.((((((.((((((((-----.(((((.((((((.....))))-))(((.((((((((.......)))))))).)))))))).)).))))))....))...)))).-- ( -45.70) >DroEre_CAF1 83706 107 + 1 ---CCCUUAC--CGCCUUCCGGUGGCCA-----UUUGCAUGCCGUCCACGAGGCG-GUGGCACAGAUUUCUGGACCCGAAGUCUGAGCCUGCGACUGAGCCGCCACUAGAAUCUGGCC-- ---.......--.((((((.((((((((-----.(((((.((((((.....))))-))(((.((((((((.(....))))))))).)))))))).)).))))))....)))...))).-- ( -45.00) >DroYak_CAF1 82139 108 + 1 ---CCCUUUC--CGCCUUCCGGUGGCCA-----UUUGCAUGCCGUCCACGAGGCGGGUGGCACAGAUUUCUGGACUCGAAGUCUGAGCCUGCGACUGAGCCAGCACUAGAAUCUGGCC-- ---.....((--((((((..((((((..-----.......)))).))..)))))))).(((.(((((((((((.(((..((((.........))))))))))).....))))))))))-- ( -41.30) >DroAna_CAF1 128434 114 + 1 ---CACUUGCCACGUUAUCCAAUGGCCACUGUAUUUGCAUGCAGUUCACG--GCU-GUGGCACAGAUUUCUGGACUUGAAGUCUGAGCCUGCGACUGAGCCACCACUAGAAUCUGGCCUU ---.....((((.(((......((((.(((((((....)))))))...((--(.(-(((((.((((((((.......)))))))).))).))).))).)))).......))).))))... ( -37.02) >consensus ___CCCUUUC__CGCCAUCCGGUGGCCA_____UUUGCAUGCCGUCCACGAGGCG_GUGGCACAGAUUUCUGGACUCGAAGUCUGAGCCUGCGACUGAGCCACCACUAGAAUCUGGCC__ .............((((((.((((((((......(((((.(((........)))....(((.((((((((.......)))))))).)))))))).)).))))))....))...))))... (-30.72 = -31.48 + 0.76)

| Location | 8,191,060 – 8,191,172 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -44.45 |
| Consensus MFE | -39.76 |
| Energy contribution | -39.45 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8191060 112 + 22224390 GCCGUCCACGAGGCG-GUGGCACAGAUUUCUGGACUCGAAGUCUGAGCCUGCGACUGAGCCACCACUAGAAUCUGGCCAAGGACUCGGCCAAAAGCUGGAGGCUGAAGGCACA (((((((....((((-(((((.(((.((.(.((.((((.....)))))).).))))).)))))).(........))))..))))((((((..........)))))).)))... ( -44.10) >DroSec_CAF1 93529 112 + 1 GCCGUCCACGAGGCG-GUGGCACAGAUUUCUGGACUCGAAGUCUGAGCCUGCGACUGAGCCACCACUAGAAUCUGGCCAAGGACUCGGCCAAAAGCUGAAGGCUGAAGGCGCA (((((((....((((-(((((.(((.((.(.((.((((.....)))))).).))))).)))))).(........))))..))))((((((..........)))))).)))... ( -44.10) >DroEre_CAF1 83736 112 + 1 GCCGUCCACGAGGCG-GUGGCACAGAUUUCUGGACCCGAAGUCUGAGCCUGCGACUGAGCCGCCACUAGAAUCUGGCCAAGGACUUGGCCAAAAGCUGAACGCUAAAGGCUGA (((.....((.((((-(((((.((((((((.(....))))))))).))).........))))))..(((....(((((((....)))))))....)))..)).....)))... ( -43.80) >DroYak_CAF1 82169 113 + 1 GCCGUCCACGAGGCGGGUGGCACAGAUUUCUGGACUCGAAGUCUGAGCCUGCGACUGAGCCAGCACUAGAAUCUGGCCAAGGACUCGGCCAAAAGCUGAAGGCUGAAGGCUGA (((((((.....((((((....((((((((.......)))))))).))))))...((.(((((.........))))))).))))((((((..........)))))).)))... ( -45.80) >consensus GCCGUCCACGAGGCG_GUGGCACAGAUUUCUGGACUCGAAGUCUGAGCCUGCGACUGAGCCACCACUAGAAUCUGGCCAAGGACUCGGCCAAAAGCUGAAGGCUGAAGGCUCA (((((((.....(((...(((.((((((((.......)))))))).))))))...((.((((...........)))))).))))((((((..........)))))).)))... (-39.76 = -39.45 + -0.31)



| Location | 8,191,060 – 8,191,172 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -44.38 |
| Consensus MFE | -40.06 |
| Energy contribution | -39.75 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8191060 112 - 22224390 UGUGCCUUCAGCCUCCAGCUUUUGGCCGAGUCCUUGGCCAGAUUCUAGUGGUGGCUCAGUCGCAGGCUCAGACUUCGAGUCCAGAAAUCUGUGCCAC-CGCCUCGUGGACGGC ..........((((((((..(((((((((....)))))))))..)).((((((((.(((((...(((((.......)))))..))...))).)))))-))).....))).))) ( -47.40) >DroSec_CAF1 93529 112 - 1 UGCGCCUUCAGCCUUCAGCUUUUGGCCGAGUCCUUGGCCAGAUUCUAGUGGUGGCUCAGUCGCAGGCUCAGACUUCGAGUCCAGAAAUCUGUGCCAC-CGCCUCGUGGACGGC ...(((.((.((....((..(((((((((....)))))))))..)).((((((((.(((((...(((((.......)))))..))...))).)))))-)))...)).)).))) ( -45.60) >DroEre_CAF1 83736 112 - 1 UCAGCCUUUAGCGUUCAGCUUUUGGCCAAGUCCUUGGCCAGAUUCUAGUGGCGGCUCAGUCGCAGGCUCAGACUUCGGGUCCAGAAAUCUGUGCCAC-CGCCUCGUGGACGGC ...........(((((((..(((((((((....)))))))))..)).(.((((((......)).(((.((((.(((.......))).)))).)))..-)))).)..))))).. ( -43.20) >DroYak_CAF1 82169 113 - 1 UCAGCCUUCAGCCUUCAGCUUUUGGCCGAGUCCUUGGCCAGAUUCUAGUGCUGGCUCAGUCGCAGGCUCAGACUUCGAGUCCAGAAAUCUGUGCCACCCGCCUCGUGGACGGC ...(((.(((((((..((..(((((((((....)))))))))..)))).)))))....(((...(((.((((.(((.......))).)))).)))...(((...))))))))) ( -41.30) >consensus UCAGCCUUCAGCCUUCAGCUUUUGGCCGAGUCCUUGGCCAGAUUCUAGUGGUGGCUCAGUCGCAGGCUCAGACUUCGAGUCCAGAAAUCUGUGCCAC_CGCCUCGUGGACGGC ...(((.((.((....((..(((((((((....)))))))))..)).(.((((((......)).(((.((((.(((.......))).)))).)))...)))).))).)).))) (-40.06 = -39.75 + -0.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:07 2006