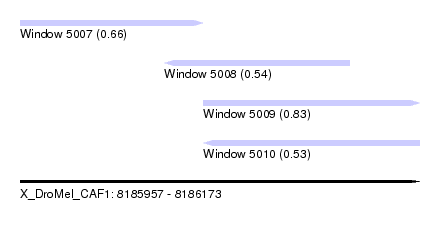
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,185,957 – 8,186,173 |
| Length | 216 |
| Max. P | 0.826073 |

| Location | 8,185,957 – 8,186,056 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -24.43 |
| Consensus MFE | -19.90 |
| Energy contribution | -19.77 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8185957 99 + 22224390 AAUGCAUACAAAAAAGUAUUCGCCAAAGGAUCCAUCGCCUUCUUGGAAAAUCAAGGGAGUGAGAGUCCUGGCAGGGCCAUUUUUUUU--UUUUGAAGGA-GC ........((((((((.....(((..((((..(.((((..(((((......)))))..)))))..)))).....))).......)))--))))).....-.. ( -24.50) >DroSec_CAF1 88616 95 + 1 AAUGCAUACAAAAAAGGAUUCGCCAAAGGAUCCAUCGCCUUCUUGGAAAAUCAAGGGAGUGAGAGUCCUGGCAGGGCCAUUUUUU-------UGAAGGAGGC ...............((((((......))))))...((((((((.(((((....((((.(....)))))(((...)))...))))-------).)))))))) ( -29.60) >DroSim_CAF1 71805 94 + 1 AAUGCAUACAAAAAAGGAUUCGCCAAAGGAUCCAUCGCCUUCUUAGA-AAUCAAGGGAGUGAGAGUCCUGGCNNNNNNNNNNNNN-------NNNNNNNNNN .....................((((..(((..(.((((.(((((.(.-...).))))))))))..))))))).............-------.......... ( -15.90) >DroEre_CAF1 78681 100 + 1 AAUGCAUACAAA-AAGGAUACGCCAAAGGAUCCGUCGCCUCCUUGGAAAAUCAAGGGAGUGGGAGUCCUGGCAGGCCCAUUUUUUUUUCUUUUGAAAAA-GC ...((.......-..((....((((..(((..(.((((..(((((......)))))..)))))..)))))))....)).....((((((....))))))-)) ( -27.70) >consensus AAUGCAUACAAAAAAGGAUUCGCCAAAGGAUCCAUCGCCUUCUUGGAAAAUCAAGGGAGUGAGAGUCCUGGCAGGGCCAUUUUUU_______UGAAGGA_GC .....................((((..(((..(.((((..(((((......)))))..)))))..))))))).............................. (-19.90 = -19.77 + -0.12)



| Location | 8,186,035 – 8,186,135 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 90.26 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -20.91 |
| Energy contribution | -21.02 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8186035 100 - 22224390 GCCAGUUCAAAGUGCCAUUUUCCAUAUUCGUUUUGGGCGCUUUGUGCCUUUGACUUGGCCCC-UCGGCCAUUGUUAAGCUGC-UCCUUCAAAA--AAAAAAAAU ((.(((.(((((((((.....(.......).....))))))))).....(((((.(((((..-..)))))..))))))))))-..........--......... ( -25.70) >DroSec_CAF1 88694 97 - 1 GCCAGUUCAAAGUGCCAUUUUCCAUAUUCGUUUUGGUCGCUUUGUGCCUUUGAGUUGGCCCCUUCGGCUAUUGUUAAGCUGCCUCCUUCA-------AAAAAAU ((((..((((((.((((....(((.........)))......)).))))))))..)))).....(((((.......))))).........-------....... ( -24.00) >DroEre_CAF1 78758 102 - 1 GCCAGUUCAAAGUGCCAUUUUCCAUAUUCGUUUUGGGCGCUUUGUGCCUUUGACUUGGCCCG-UCGGCCAUUGUUAAGCUGC-UUUUUCAAAAGAAAAAAAAAU ((.(((.(((((((((.....(.......).....))))))))).....(((((.(((((..-..)))))..))))))))))-((((((....))))))..... ( -28.00) >consensus GCCAGUUCAAAGUGCCAUUUUCCAUAUUCGUUUUGGGCGCUUUGUGCCUUUGACUUGGCCCC_UCGGCCAUUGUUAAGCUGC_UCCUUCAAAA__AAAAAAAAU ((..((.((((((((((................))).))))))).))..(((((.(((((.....)))))..)))))))......................... (-20.91 = -21.02 + 0.11)


| Location | 8,186,056 – 8,186,173 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.75 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -26.41 |
| Energy contribution | -26.73 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
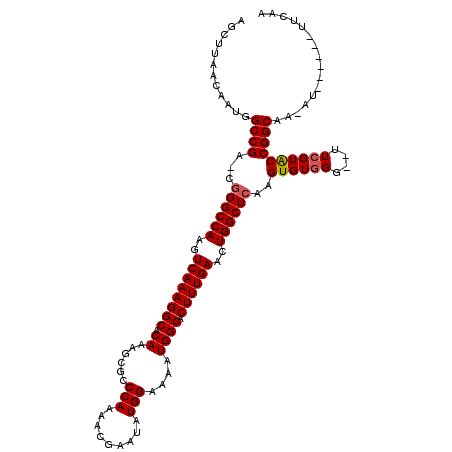
>X_DroMel_CAF1 8186056 117 + 22224390 AGCUUAACAAUGGCCGA-GGGGCCAAGUCAAAGGCACAAAGCGCCCAAAACGAAUAUGGAAAAUGGCACUUUGAACUGGCUCAAGUGUGGG--UUCGUACCGGCAAAAUGCAAAAUGCAA ............(((..-.((((((..((((((((.....))(((.....(......)......))).))))))..)))))).......((--(....))))))....(((.....))). ( -32.20) >DroSec_CAF1 88711 114 + 1 AGCUUAACAAUAGCCGAAGGGGCCAACUCAAAGGCACAAAGCGACCAAAACGAAUAUGGAAAAUGGCACUUUGAACUGGCUCAAGUGUGGGUGUUCGUACCGGCGAAAU------UUCAA .(((.......))).((((((((((..((((((((.((......(((.........)))....)))).))))))..))))))...(((.((((....)))).)))...)------))).. ( -29.60) >DroEre_CAF1 78781 110 + 1 AGCUUAACAAUGGCCGA-CGGGCCAAGUCAAAGGCACAAAGCGCCCAAAACGAAUAUGGAAAAUGGCACUUUGAACUGGCUGAAGUGUGGG--UUCGUGCCGGCAA-AU------UUCAA .(((......(((((..-..))))).......(((((((((.(((.....(......)......))).))))(((((.((......)).))--)))))))))))..-..------..... ( -35.70) >DroYak_CAF1 76681 110 + 1 AGCUUAACAAUGGCCGA-CGGGCCAAGUCAAAGGCACAAAGCGGCCAAAACGAAUAUGGAAAAUGGCACUUUGAACUGGCUCAAGUGUUGG--UUCGUACCGGCAA-AU------UUCAU .(((.......)))...-.((((((..((((((((.....)).((((................)))).))))))..))))))...((((((--(....))))))).-..------..... ( -32.49) >consensus AGCUUAACAAUGGCCGA_CGGGCCAAGUCAAAGGCACAAAGCGCCCAAAACGAAUAUGGAAAAUGGCACUUUGAACUGGCUCAAGUGUGGG__UUCGUACCGGCAA_AU______UUCAA ............((((...((((((..((((((((.((......(((.........)))....)))).))))))..))))))..((((((....))))))))))................ (-26.41 = -26.73 + 0.31)


| Location | 8,186,056 – 8,186,173 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.75 |
| Mean single sequence MFE | -30.54 |
| Consensus MFE | -26.01 |
| Energy contribution | -26.14 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
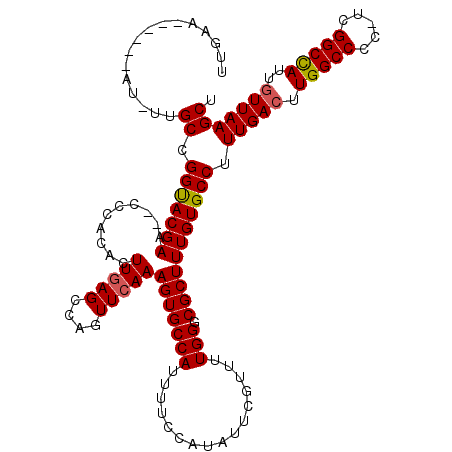
>X_DroMel_CAF1 8186056 117 - 22224390 UUGCAUUUUGCAUUUUGCCGGUACGAA--CCCACACUUGAGCCAGUUCAAAGUGCCAUUUUCCAUAUUCGUUUUGGGCGCUUUGUGCCUUUGACUUGGCCCC-UCGGCCAUUGUUAAGCU .(((.....)))....((.((((((((--(..............))))((((((((.....(.......).....))))))))))))).(((((.(((((..-..)))))..))))))). ( -32.84) >DroSec_CAF1 88711 114 - 1 UUGAA------AUUUCGCCGGUACGAACACCCACACUUGAGCCAGUUCAAAGUGCCAUUUUCCAUAUUCGUUUUGGUCGCUUUGUGCCUUUGAGUUGGCCCCUUCGGCUAUUGUUAAGCU .....------.....((((((......))).........((((..((((((.((((....(((.........)))......)).))))))))..))))......)))............ ( -27.20) >DroEre_CAF1 78781 110 - 1 UUGAA------AU-UUGCCGGCACGAA--CCCACACUUCAGCCAGUUCAAAGUGCCAUUUUCCAUAUUCGUUUUGGGCGCUUUGUGCCUUUGACUUGGCCCG-UCGGCCAUUGUUAAGCU .....------..-..((.((((((((--(..............))))((((((((.....(.......).....))))))))))))).(((((.(((((..-..)))))..))))))). ( -32.84) >DroYak_CAF1 76681 110 - 1 AUGAA------AU-UUGCCGGUACGAA--CCAACACUUGAGCCAGUUCAAAGUGCCAUUUUCCAUAUUCGUUUUGGCCGCUUUGUGCCUUUGACUUGGCCCG-UCGGCCAUUGUUAAGCU .....------..-..((.((((((((--.(.....(((((....))))).(.((((................)))))).)))))))).(((((.(((((..-..)))))..))))))). ( -29.29) >consensus UUGAA______AU_UUGCCGGUACGAA__CCCACACUUGAGCCAGUUCAAAGUGCCAUUUUCCAUAUUCGUUUUGGGCGCUUUGUGCCUUUGACUUGGCCCC_UCGGCCAUUGUUAAGCU ................((.(((((((..........(((((....)))))(((((((................))).))))))))))).(((((.(((((.....)))))..))))))). (-26.01 = -26.14 + 0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:04 2006