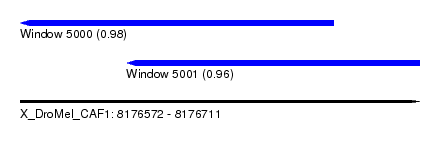
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,176,572 – 8,176,711 |
| Length | 139 |
| Max. P | 0.982815 |

| Location | 8,176,572 – 8,176,681 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.69 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -18.84 |
| Energy contribution | -19.32 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8176572 109 - 22224390 -------AAAGCCAAGCCCACCCCACCGCCGUAUACCCUUCAUUUUACUCAAGUGGCGACUUAAAGUCGCCCUAAUUGAAUUGCGCAAUAAUGUAGCAACUU-UUCAGGGGCACUGA -------........((((................................((.((((((.....))))))))..((((((((((((....))).))))...-)))))))))..... ( -26.60) >DroSec_CAF1 79871 110 - 1 -------AGAAGCCACCCCACCCCACCACCGUAUACCCUUCAUUUUACUCAAGUGGCGACUUAAAGUCGCCCUAAUUGAAUUGCGCAAUAAUGUAGCAACUUUUUCUGGGGCACUGA -------.............(((((.......................((((..((((((.....))))))....)))).(((((((....))).)))).......)))))...... ( -25.00) >DroSim_CAF1 62270 110 - 1 -------AAAAGCCACCCCACCCCACCAUCGUAUACCCUUCAUUUUACUCAAGUGGCGACUUAAAGUCGCCCUAAUUGAAUUGCGCAAUAAUGUAGCAACUUUUUCAGGGGCACUGA -------.............((((........................((((..((((((.....))))))....)))).(((((((....))).))))........))))...... ( -24.70) >DroEre_CAF1 69963 99 - 1 CCUCAC----------CAGCCC-------CAUACACCCUUCAUUUUACUCAAGUGGCGACUUAAAGUCGCCCUAAUUGAAUUGCGCAAUAAUGUCGCAACUU-UUCAGGGGCACUGA ......----------..((((-------(.....................((.((((((.....))))))))...(((((((((((....)).)))))...-)))))))))..... ( -31.30) >DroYak_CAF1 67717 116 - 1 CCUCAACACACCCCACCACACCCCCCCUUUAUACACCCUUCAUUUUACUCAAGUGGCGACUUAAAGUCGCCCUAAUUGAAUUGCGCAAUAAUGUAGCAACUU-UUCAGGGGCACUGA ..........((((.....................................((.((((((.....))))))))...(((((((((((....))).))))...-))))))))...... ( -25.00) >DroAna_CAF1 109015 96 - 1 ------------CCGCCCCAC-----UCGGGCCAACCACCCACCUCCCCCAAGUGGCGACUUAAAGUCUCCCUAAUUGAAUUGCGCAAUAAUGUGACAACUU-U-GGGGGGC--AAA ------------..((((...-----..))))............(((((((((.((.(((.....))).)))).......(((((((....)))).)))...-)-)))))).--... ( -28.90) >consensus _______A_A__CCACCCCACCCCACCACCGUAUACCCUUCAUUUUACUCAAGUGGCGACUUAAAGUCGCCCUAAUUGAAUUGCGCAAUAAUGUAGCAACUU_UUCAGGGGCACUGA ...................................(((((........((((..((((((.....))))))....)))).(((((((....))).)))).......)))))...... (-18.84 = -19.32 + 0.47)



| Location | 8,176,609 – 8,176,711 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 75.16 |
| Mean single sequence MFE | -17.98 |
| Consensus MFE | -13.75 |
| Energy contribution | -13.63 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8176609 102 - 22224390 ACCCCUCAACGUCUCCCACCUUUGGGUGAA-AAAGCCAAGCCCACCCCACCGCCGUAUACCCUUCAUUUUACUCAAGUGGCGACUUAAAGUCGCCCUAAUUGA .....((((..((.((((....)))).)).-............................................((.((((((.....))))))))..)))) ( -19.10) >DroSec_CAF1 79909 101 - 1 AACC-CCAACGUCUCCCACCUUUGGGCCAA-AGAAGCCACCCCACCCCACCACCGUAUACCCUUCAUUUUACUCAAGUGGCGACUUAAAGUCGCCCUAAUUGA ...(-((((.(........).))))).(((-.((((.........................))))..........((.((((((.....))))))))..))). ( -17.41) >DroSim_CAF1 62308 101 - 1 AACC-CCAACGUCUCCCACCUUUGGGCCAA-AAAAGCCACCCCACCCCACCAUCGUAUACCCUUCAUUUUACUCAAGUGGCGACUUAAAGUCGCCCUAAUUGA ...(-((((.(........).)))))....-.........................................((((..((((((.....))))))....)))) ( -16.80) >DroAna_CAF1 109049 91 - 1 GGCC-CCUCCCCCCCGCACCU-UGUGCAUUA-----CCGCCCCAC-----UCGGGCCAACCACCCACCUCCCCCAAGUGGCGACUUAAAGUCUCCCUAAUUGA ((((-(.........((((..-.))))....-----..(.....)-----..)))))..................((.((.(((.....))).))))...... ( -18.60) >consensus AACC_CCAACGUCUCCCACCUUUGGGCCAA_AAAAGCCACCCCACCCCACCACCGUAUACCCUUCAUUUUACUCAAGUGGCGACUUAAAGUCGCCCUAAUUGA ..............((((....)))).................................................((.((((((.....))))))))...... (-13.75 = -13.63 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:56 2006