
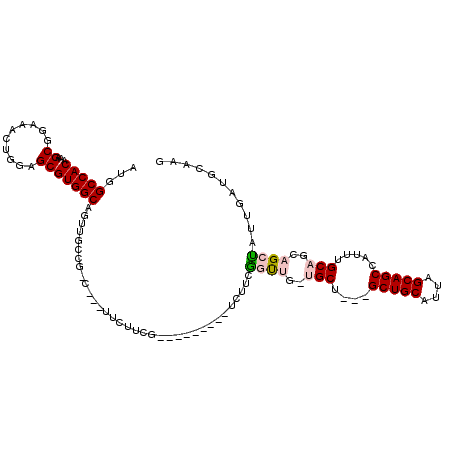
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,171,795 – 8,171,921 |
| Length | 126 |
| Max. P | 0.990206 |

| Location | 8,171,795 – 8,171,889 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.79 |
| Mean single sequence MFE | -36.42 |
| Consensus MFE | -20.86 |
| Energy contribution | -21.20 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8171795 94 + 22224390 AUGGCCACAAAGCGGAAAGUGGAGCGUGGCUGG----------UUCUUCG---------UCUUCGGCUGAUGCA---GCUGCAUUAGCAGCCAUUUGCAACAGCCAUUGAUGGCAG ...(((((((.(.(((....(((((.......)----------))))...---------))).)(((((.((((---(((((....)))))....)))).))))).))).)))).. ( -36.40) >DroVir_CAF1 65114 111 + 1 AUGGCCACAAAGCGGAAACUGGAGCGUGGCAACUGACGAC---GACGUUG-UAAAAAUUGCUGCAGUUGUUGCUGCUGCUGCAUUAGCAGCCAUUUGCAGCAGCUAUUGAUGCGC- ...((((((((((((.(((((.((((..(((((.......---...))))-)......)))).))))).)))))(((((((((............)))))))))..))).)).))- ( -38.80) >DroPse_CAF1 76662 97 + 1 AUGGCCACAAAGCGGAAACUGGAGCGUGGCAGUUGCUG-C---UUCUUCA---------UCUUUGCUC---GCU---GCUGCAUUAGCAGCCAUUUGCAGCAGCUAUUGAUGUAAG ...(((((.....(....)......)))))((((((((-(---.....((---------....))...---(((---((((...))))))).....)))))))))........... ( -31.90) >DroMoj_CAF1 80245 108 + 1 AUGGCCACAAAGCGGAAACUGGAGCGUGGCAACUGACGGC---GACGUUG-UAAAAAUUGCUGCAGUUGUUGCU---GCUGCAUUAGCAGCCAUUUGCAGCAGCUAUUGAUGCGU- ...((..(((((((....)((.(((((.((........))---.))))).-)).....((((((((.((..(((---((((...))))))))).))))))))))).)))..))..- ( -38.40) >DroAna_CAF1 103057 112 + 1 AUGGCCACAAAGCGGAAACUGGAGCGUGGCAGUUGCAG-CUUCUUCUUCGCCCUCGUCUUCUUCGGCUGAUGCA---GCUGCAUUAGCAGCCAUUUGCAGAAGCCAUUGAUGCCAC .((((..(((.(((((....(((((...((....)).)-))))...))))).............((((..((((---(((((....)))))....))))..)))).)))..)))). ( -41.10) >DroPer_CAF1 76558 97 + 1 AUGGCCACAAAGCGGAAACUGGAGCGUGGCAGUUGCUG-C---UUCUUCA---------UCUUUGCUC---GCU---GCUGCAUUAGCAGCCAUUUGCAGCAGCUAUUGAUGUAAG ...(((((.....(....)......)))))((((((((-(---.....((---------....))...---(((---((((...))))))).....)))))))))........... ( -31.90) >consensus AUGGCCACAAAGCGGAAACUGGAGCGUGGCAGUUGCCG_C___UUCUUCG_________UCUUCGGUUG_UGCU___GCUGCAUUAGCAGCCAUUUGCAGCAGCUAUUGAUGCAAG ...(((((...((..........)))))))..................................((((..(((....(((((....))))).....)))..))))........... (-20.86 = -21.20 + 0.34)

| Location | 8,171,828 – 8,171,921 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.98 |
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -18.44 |
| Energy contribution | -19.83 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8171828 93 + 22224390 ----UUCUUCG---------UCUUCGGCUGAUGCAGCUGCAUUAGCAGCCAUUUGCAACAGCCAUUGAUGGCAGUGCAUAUGUGUUUGUU-GCUCUUCCCUCUCUCU- ----.......---------.....(((((.(((((((((....)))))....)))).)))))...((.((.((.(((...........)-)).)).)).)).....- ( -29.30) >DroSec_CAF1 75082 93 + 1 ----UUCUUCG---------UCUUCGGCUGAUGCAGCUGCAUUAGCAGCCAUUUGCAACAGCCAUUGAUGGCAGUGCAUAUGUGUUUGUU-GCUCUUACCUCUCGCU- ----.....((---------((...(((((.(((((((((....)))))....)))).)))))...))))((((.(((....)))...))-))..............- ( -29.00) >DroEre_CAF1 64830 94 + 1 ----UUCUUCG---------UCUUCGGCUGAUGCAGCUGCAUUAGCAGCCAUUUGCAACAGCCAUUGAUGGCGGUGCAUAUGUGUUUGUU-GCUCGUCCCUCUCUCUU ----.....((---------((...(((((.(((((((((....)))))....)))).)))))...))))((((.(((...........)-))))))........... ( -30.20) >DroYak_CAF1 62864 93 + 1 ----UUCUUCG---------UCUUCGGCUGAUGCAGCUGCAUUAGCAGCCAUUUGCAACAGCCAUUGAUGGCGGUGCAUAUGUGUUUGUU-GCUCUGCUCUCUCUCU- ----.....((---------((...(((((.(((((((((....)))))....)))).)))))...))))((((.(((...........)-)).)))).........- ( -33.90) >DroAna_CAF1 103095 93 + 1 CUUCUUCUUCGCCCUCGUCUUCUUCGGCUGAUGCAGCUGCAUUAGCAGCCAUUUGCAGAAGCCAUUGAUGCCACUGCAUAUGUAUCUCUC-CCU-------------- ..........(((............))).(((((((((((....)))))....(((((..((.......))..)))))..))))))....-...-------------- ( -23.60) >DroPer_CAF1 76596 80 + 1 C---UUCUUCA---------UCUUUGCUC---GCUGCUGCAUUAGCAGCCAUUUGCAGCAGCUAUUGAUGUAAGUGCAUAUGUGUUUGUAUGCAU------------- .---..(((((---------((..((...---(((((((((............)))))))))))..)))).)))(((((((......))))))).------------- ( -27.20) >consensus ____UUCUUCG_________UCUUCGGCUGAUGCAGCUGCAUUAGCAGCCAUUUGCAACAGCCAUUGAUGGCAGUGCAUAUGUGUUUGUU_GCUCUUCCCUCUCUCU_ ..........................((((((((....))))))))((((((.((((...((((....))))..)))).))).)))...................... (-18.44 = -19.83 + 1.39)


| Location | 8,171,828 – 8,171,921 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 77.98 |
| Mean single sequence MFE | -27.83 |
| Consensus MFE | -18.58 |
| Energy contribution | -19.47 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8171828 93 - 22224390 -AGAGAGAGGGAAGAGC-AACAAACACAUAUGCACUGCCAUCAAUGGCUGUUGCAAAUGGCUGCUAAUGCAGCUGCAUCAGCCGAAGA---------CGAAGAA---- -.....((.((.((.((-(...........))).)).)).))..((((((.((((....(((((....))))))))).))))))....---------.......---- ( -29.10) >DroSec_CAF1 75082 93 - 1 -AGCGAGAGGUAAGAGC-AACAAACACAUAUGCACUGCCAUCAAUGGCUGUUGCAAAUGGCUGCUAAUGCAGCUGCAUCAGCCGAAGA---------CGAAGAA---- -..((.((((((.(.((-(...........))).))))).))..((((((.((((....(((((....))))))))).))))))....---------)).....---- ( -30.00) >DroEre_CAF1 64830 94 - 1 AAGAGAGAGGGACGAGC-AACAAACACAUAUGCACCGCCAUCAAUGGCUGUUGCAAAUGGCUGCUAAUGCAGCUGCAUCAGCCGAAGA---------CGAAGAA---- ......((.((.((.((-(...........)))..)))).))..((((((.((((....(((((....))))))))).))))))....---------.......---- ( -28.30) >DroYak_CAF1 62864 93 - 1 -AGAGAGAGAGCAGAGC-AACAAACACAUAUGCACCGCCAUCAAUGGCUGUUGCAAAUGGCUGCUAAUGCAGCUGCAUCAGCCGAAGA---------CGAAGAA---- -.....((..((.(.((-(...........))).).))..))..((((((.((((....(((((....))))))))).))))))....---------.......---- ( -28.20) >DroAna_CAF1 103095 93 - 1 --------------AGG-GAGAGAUACAUAUGCAGUGGCAUCAAUGGCUUCUGCAAAUGGCUGCUAAUGCAGCUGCAUCAGCCGAAGAAGACGAGGGCGAAGAAGAAG --------------...-....(((.(((.(((((.(((.......))).))))).)))(((((....)))))...))).(((............))).......... ( -26.30) >DroPer_CAF1 76596 80 - 1 -------------AUGCAUACAAACACAUAUGCACUUACAUCAAUAGCUGCUGCAAAUGGCUGCUAAUGCAGCAGC---GAGCAAAGA---------UGAAGAA---G -------------.((((((........))))))(((.((((....(((((((((...(....)...)))))))))---.......))---------)))))..---. ( -25.10) >consensus _AGAGAGAGGGAAGAGC_AACAAACACAUAUGCACUGCCAUCAAUGGCUGUUGCAAAUGGCUGCUAAUGCAGCUGCAUCAGCCGAAGA_________CGAAGAA____ ............................................((((((.((((....(((((....))))))))).))))))........................ (-18.58 = -19.47 + 0.89)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:53 2006