| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,168,725 – 8,168,816 |
| Length | 91 |
| Max. P | 0.991574 |

| Location | 8,168,725 – 8,168,816 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 72.80 |
| Mean single sequence MFE | -22.00 |
| Consensus MFE | -10.90 |
| Energy contribution | -11.93 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.50 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8168725 91 - 22224390 -----------UGAGUGAAAGUGCUGGCGAAAUUAUUGACGACUGCAUUUUCGGGCUGUAAAUCAAGCAAAAGUGAAAAA---AAGUAAAAAAAAAAAUUGGAAU -----------....(((((((((...((..........))...))))))))).(((........)))............---...................... ( -13.40) >DroPse_CAF1 73746 85 - 1 -----------GGAGAGAAAGUGCUGGGAAAAUUAUUGCAGACUGCAUCUCUGCGCUGUAAAUCAAGCGAAAGUGAAAACCUCCAGCAAAAA-AAAA-------- -----------(.(((((.(((.(((.((......)).))))))...))))).)((((........((....)).........)))).....-....-------- ( -21.53) >DroSec_CAF1 72088 90 - 1 --------CUCUGAGUGAAAGUGCUGGCGAAAUUAUUGCCGCCUGCAUUUUCGGGCUGUAAAUCAAGCGAAAGUGGAAA----UAGUACAAAAAA---GUGGAAU --------.(((....((((((((.((((..........)))).))))))))(..((((...((..((....)).)).)----)))..)......---..))).. ( -24.10) >DroSim_CAF1 54123 92 - 1 --------UUCUGAGUGAAAGUGCUGGCGAAAUUAUUGCCGCCUGCAUUUUCGGGCUGUAAAUCAAGCGAAAGUGAAAA----AAGUACAAAAAA-ACGAGGAAU --------((((..((((((((((.((((..........)))).))))))))....((((......((....)).....----...)))).....-))..)))). ( -25.74) >DroYak_CAF1 59648 91 - 1 UGAAAGUGCACCGAGUGAAAGUGCUGGCAAAAUUAUUGCCGACUGCAUUUUCGGGCUGUAAGUCAAGCGAAAGUACAAA-------------AAC-CGAAAGAAC (((...((((((...(((((((((((((((.....))))))...))))))))))).))))..))).((....)).....-------------...-(....)... ( -25.70) >DroPer_CAF1 73649 86 - 1 -----------GGAGAGAAAGUGCUGGGAAAAUUAUUGCAGACUGCAUCUCUGCGCUGUAAAUCAAGCGAAAGUGAAAACCUCCAGCAAAAAAAAAA-------- -----------(.(((((.(((.(((.((......)).))))))...))))).)((((........((....)).........))))..........-------- ( -21.53) >consensus ___________UGAGUGAAAGUGCUGGCAAAAUUAUUGCCGACUGCAUUUUCGGGCUGUAAAUCAAGCGAAAGUGAAAA____AAGUAAAAAAAA_A___GGAAU ................((((((((((((((.....))))))...))))))))..(((........)))..................................... (-10.90 = -11.93 + 1.03)

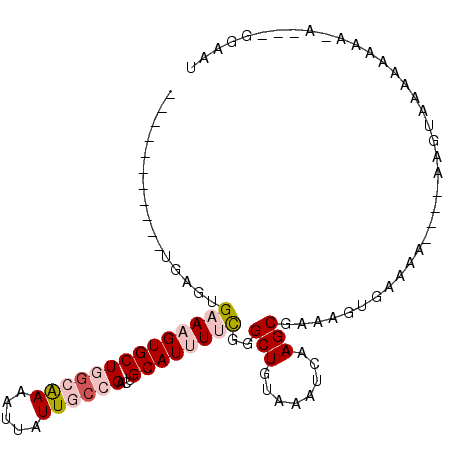
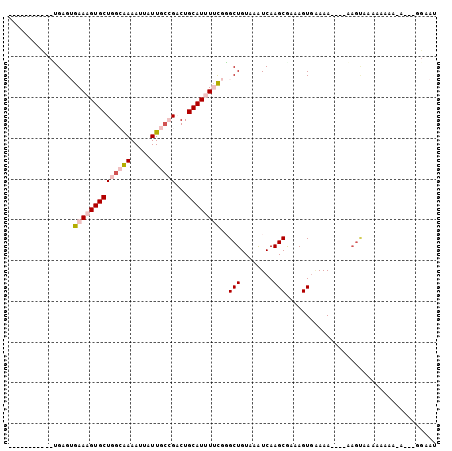
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:48 2006