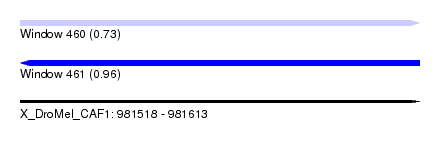
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 981,518 – 981,613 |
| Length | 95 |
| Max. P | 0.960454 |

| Location | 981,518 – 981,613 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 84.35 |
| Mean single sequence MFE | -12.07 |
| Consensus MFE | -8.62 |
| Energy contribution | -9.12 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 981518 95 + 22224390 AAUGAAAAUGAAGUUGCCUUGGGCAACUUUCCCCCUCCUUCUAGCCCCCCCUU-AAUAAAAAAUAAGAAAUAAAAAAGGAAAACCCAAAAACAAAA .........(((((((((...))))))))).....(((((..........(((-(........))))........)))))................ ( -13.57) >DroSec_CAF1 64273 85 + 1 AAUGAAAAUUAAGUUGCCUUGGGCAACUUUCCCCC---UUCUGGCC--CCCUU-AAUAAAAAAUAAGAAAUAAAAAAGGAAAACCCAAAAA----- ............((((((...))))))(((((.((---....))..--..(((-(........))))..........))))).........----- ( -12.70) >DroSim_CAF1 43495 85 + 1 AAUGAAAAUGAAGUUGCCUUGGGCAACUUUCCCCC---UUCUGGCC--CCCUU-AAUAAAAAAUAAGAAAUAAAAAAGGAAAACCCAAAAA----- .........(((((((((...)))))))))...((---((......--..(((-(........))))........))))............----- ( -13.29) >DroYak_CAF1 69827 82 + 1 AAUGAAAAUGAAGGUGCCUUGGGCAACUUUCCCCC---UUCUAGCC--CCCUUGAAUAAAAAAACUAAAAUGGAAAAA----AAAUAAAAA----- .........(((((((((...))).))))))....---(((((...--......................)))))...----.........----- ( -8.71) >consensus AAUGAAAAUGAAGUUGCCUUGGGCAACUUUCCCCC___UUCUAGCC__CCCUU_AAUAAAAAAUAAGAAAUAAAAAAGGAAAACCCAAAAA_____ .........(((((((((...))))))))).................................................................. ( -8.62 = -9.12 + 0.50)

| Location | 981,518 – 981,613 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 84.35 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -14.35 |
| Energy contribution | -14.97 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 981518 95 - 22224390 UUUUGUUUUUGGGUUUUCCUUUUUUAUUUCUUAUUUUUUAUU-AAGGGGGGGCUAGAAGGAGGGGGAAAGUUGCCCAAGGCAACUUCAUUUUCAUU ..((.(((((((.((..((((.....................-))))..)).))))))).))(..((((((((((...))))))))..))..)... ( -24.70) >DroSec_CAF1 64273 85 - 1 -----UUUUUGGGUUUUCCUUUUUUAUUUCUUAUUUUUUAUU-AAGGG--GGCCAGAA---GGGGGAAAGUUGCCCAAGGCAACUUAAUUUUCAUU -----....((((((..((((((...(..((((........)-)))..--)...))))---))..))((((((((...))))))))....)))).. ( -24.30) >DroSim_CAF1 43495 85 - 1 -----UUUUUGGGUUUUCCUUUUUUAUUUCUUAUUUUUUAUU-AAGGG--GGCCAGAA---GGGGGAAAGUUGCCCAAGGCAACUUCAUUUUCAUU -----....((((((..((((((...(..((((........)-)))..--)...))))---))..))((((((((...))))))))....)))).. ( -23.70) >DroYak_CAF1 69827 82 - 1 -----UUUUUAUUU----UUUUUCCAUUUUAGUUUUUUUAUUCAAGGG--GGCUAGAA---GGGGGAAAGUUGCCCAAGGCACCUUCAUUUUCAUU -----.......((----(((..((.(((((((((((((....)))))--))))))))---))..))))).((((...)))).............. ( -19.30) >consensus _____UUUUUGGGUUUUCCUUUUUUAUUUCUUAUUUUUUAUU_AAGGG__GGCCAGAA___GGGGGAAAGUUGCCCAAGGCAACUUCAUUUUCAUU ......((((((.(..(((((...((............))...)))))..).))))))....(..((((((((((...))))))))..))..)... (-14.35 = -14.97 + 0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:15 2006